Parallel Looping & Variants
Daniel Anderson
Week 5
Learning objectives
Understand the differences between
map,map2, andpmapKnow when to apply
walkinstead ofmap, and why it may be usefulUnderstand the similarities and differences between
mapandmodifyDiagnose errors with
safelyand understand other situations where it may be helpfulCollapsing/reducing lists with
purrr::reduce()orbase::Reduce()
Simulate!
sim1 <- map2(conditions$n, conditions$mu, ~{ rnorm(n = .x, mean = .y, sd = 10)})str(sim1)## List of 510## $ : num [1:5] -2.135 10.466 11.083 -0.149 18.545## $ : num [1:10] -8.72 -13.05 1.54 6.27 -7.47 ...## $ : num [1:15] 11.1 -16.15 -10.05 4.72 10.5 ...## $ : num [1:20] 0.227 8.498 -20.939 -22.962 -0.948 ...## $ : num [1:25] 15.86 -7.01 -5.43 -1.81 -5.86 ...## $ : num [1:30] -1.54 5.95 -0.14 13.83 6.79 ...## $ : num [1:35] -0.588 6.627 -12.193 2.372 -13.161 ...## $ : num [1:40] -5.34 7.05 -2.53 12.09 4.39 ...## $ : num [1:45] -7.43 -1.1 -9.65 -8.73 -1.87 ...## $ : num [1:50] -11.61 -5.59 -2.1 -17.48 -6.56 ...## $ : num [1:55] -8.05 -2.02 13.7 -14.15 -6.81 ...## $ : num [1:60] -5.527 -12.283 -13.94 6.904 -0.334 ...## $ : num [1:65] -6.2518 -1.5114 -0.0274 3.5003 -9.0024 ...## $ : num [1:70] 15.294 -8.09 0.538 -7.834 -7.617 ...## $ : num [1:75] -13.55 0.984 -0.64 -11.444 -3.83 ...## $ : num [1:80] 11.25 8.55 8.89 -19.8 5.67 ...## $ : num [1:85] -2.35 -5.68 16.43 4.82 12.68 ...## $ : num [1:90] -19.095 13.054 -15.354 -0.498 -7.509 ...## $ : num [1:95] 9.06 2.6 -17.42 12.68 -13.58 ...## $ : num [1:100] 14.78 -2.65 14.25 -5.68 1.08 ...## $ : num [1:105] -1.44 -12.11 12.62 -3.27 2.44 ...## $ : num [1:110] -16.27 7.98 -5.97 -2.8 -14.41 ...## $ : num [1:115] 9.54 16.85 -7.4 -11.38 -1.09 ...## $ : num [1:120] 5.31 6.976 5.66 -6.599 -0.844 ...## $ : num [1:125] 4.66 -12.18 11.78 -6.08 -2.37 ...## $ : num [1:130] 1.419 1.797 -0.556 2.156 -14.721 ...## $ : num [1:135] -0.7536 0.0431 -11.0482 -21.7445 4.5122 ...## $ : num [1:140] -5.091 -0.662 8.399 9.693 -2.69 ...## $ : num [1:145] -4.47 12.38 -9.18 -18.92 -25.53 ...## $ : num [1:150] -12.57 6.89 -3.88 -13.99 -9.94 ...## $ : num [1:5] 13.25 8.59 -19.07 -2.13 -4.46## $ : num [1:10] -11.118 -0.335 8.384 10.266 -10.506 ...## $ : num [1:15] -13.99 -9.18 -3.12 2.55 -16.86 ...## $ : num [1:20] 0.486 15.647 9.483 10.967 -5.976 ...## $ : num [1:25] -5.69 6.26 4.24 -5.56 23.39 ...## $ : num [1:30] -14.364 -0.686 -9.779 3.741 -3.284 ...## $ : num [1:35] -21.15 16.41 -5.03 -5.13 -10.06 ...## $ : num [1:40] 4.42 3.28 4.11 -5.49 -12.07 ...## $ : num [1:45] -6.59 15.53 21.61 9.07 -3.76 ...## $ : num [1:50] -0.0784 9.1196 -19.4697 -7.6971 4.5796 ...## $ : num [1:55] 0.378 -0.779 1.344 -5.242 -9.46 ...## $ : num [1:60] -2.39 -13.4 4.41 -11.93 3.71 ...## $ : num [1:65] 19.48 -14.76 -5.83 9.05 18.81 ...## $ : num [1:70] 26.3 -2.95 -5.87 -5.42 4.98 ...## $ : num [1:75] 21.731 -4.778 2.62 0.653 -1.628 ...## $ : num [1:80] 12 8.24 2.81 12.06 2.06 ...## $ : num [1:85] -1.122 0.697 -17.395 3.086 -8.11 ...## $ : num [1:90] -12.17 -3.49 6.28 -4.22 10.07 ...## $ : num [1:95] -11.992 -2.839 -0.541 -9.832 5.252 ...## $ : num [1:100] -10.18 -5.09 8.5 4.57 6.16 ...## $ : num [1:105] -8.77 12.529 -5.545 -0.231 3.304 ...## $ : num [1:110] -4.64 5.71 2.79 -4.5 -7.36 ...## $ : num [1:115] -5.25 -18.23 -5.65 2.56 -5.28 ...## $ : num [1:120] 3.254 2.861 -3.972 -15.111 -0.886 ...## $ : num [1:125] 2.09 -6.01 6.15 4.5 -4.73 ...## $ : num [1:130] -5.05 -9.92 6.77 -11.94 -7.53 ...## $ : num [1:135] 3.399 -0.886 -15.508 13.685 -10.01 ...## $ : num [1:140] 9.36 9.33 6.97 -5.94 23.07 ...## $ : num [1:145] -3.5 -5.47 2.14 6.78 0.89 ...## $ : num [1:150] -4.93 -6.83 -1.33 -4.82 -10.52 ...## $ : num [1:5] 3.4 -4.65 -6.8 -13.84 7## $ : num [1:10] 8.878 -12.646 0.865 -6.316 -6.799 ...## $ : num [1:15] 0.199 -16.621 -6.265 5.45 -13.263 ...## $ : num [1:20] 8.3333 16.9791 -12.3281 13.4383 -0.0692 ...## $ : num [1:25] -6.42 4.34 -6.42 15.24 4.83 ...## $ : num [1:30] 2.28 -15.83 -9.63 -9.63 -3.59 ...## $ : num [1:35] -2.99 -1.61 -5.34 14.5 25.4 ...## $ : num [1:40] -6.3019 1.1355 2.9021 0.0776 -10.4499 ...## $ : num [1:45] -5.75 -3.33 16.42 13.11 -8.82 ...## $ : num [1:50] -7.48 -11.52 17.08 -4.13 -17.37 ...## $ : num [1:55] -11 1.06 3.13 3.25 -15.81 ...## $ : num [1:60] -7.699 -8.987 -9.756 -4.217 0.813 ...## $ : num [1:65] -19.05 -29.28 -4.01 18.1 -3.92 ...## $ : num [1:70] -0.89 -6.65 -2.4 -10.28 -5.57 ...## $ : num [1:75] 0.164 15.412 1.634 -10.484 3.918 ...## $ : num [1:80] 6.47 2.44 3.81 -5.79 17.68 ...## $ : num [1:85] -0.828 12.719 6.7 4.086 -3.096 ...## $ : num [1:90] -7.042 15.799 -13.421 4.396 -0.536 ...## $ : num [1:95] -6.5 -2.18 13.79 3.09 -21.64 ...## $ : num [1:100] -17.38 1.917 -0.516 1.215 -8.002 ...## $ : num [1:105] -2.232 -0.515 17.965 -18.168 8.545 ...## $ : num [1:110] 13.96 0.428 -1.474 -2.781 5.572 ...## $ : num [1:115] -9.82 -2.89 3.26 -1.94 -17.93 ...## $ : num [1:120] -20.51 2.18 -13.14 7.23 -13.92 ...## $ : num [1:125] -5.66 -11.54 -7.89 -2.98 -12.02 ...## $ : num [1:130] -2.61 -18.4 8.95 12.24 -13.42 ...## $ : num [1:135] -7.1 -3.62 -14.76 -2.02 17.41 ...## $ : num [1:140] 7.12 9.5 5.29 4.47 7.26 ...## $ : num [1:145] -0.477 -15.662 -2.699 0.856 -22.186 ...## $ : num [1:150] 7.025 4.514 0.778 -5.803 9.802 ...## $ : num [1:5] -8.17 -12.21 -1.71 2.84 -6.24## $ : num [1:10] 6.98 3.8 -5.07 -3.84 4.11 ...## $ : num [1:15] 19.37 -5.85 2.29 -3.47 10.2 ...## $ : num [1:20] -2.4492 -5.8553 15.8816 0.1707 -0.0201 ...## $ : num [1:25] -8.48 -6.87 -4.92 18.76 -6.2 ...## $ : num [1:30] 11.588 -18.958 1.722 -0.312 10.077 ...## $ : num [1:35] -16.25 -2.3 -4.82 -9.78 -7.88 ...## $ : num [1:40] 1.89 1.28 -2.77 -4.49 1.48 ...## $ : num [1:45] 10.27 18.17 2.79 -5.57 -2.35 ...## [list output truncated]More powerful
Add it as a list column!
sim2 <- conditions %>% as_tibble() %>% # Not required, but definitely helpful mutate(sim = map2(n, mu, ~rnorm(n = .x, mean = .y, sd = 10))) sim2## # A tibble: 510 × 3## n mu sim ## <dbl> <dbl> <list> ## 1 5 -2 <dbl [5]> ## 2 10 -2 <dbl [10]>## 3 15 -2 <dbl [15]>## 4 20 -2 <dbl [20]>## 5 25 -2 <dbl [25]>## 6 30 -2 <dbl [30]>## 7 35 -2 <dbl [35]>## 8 40 -2 <dbl [40]>## 9 45 -2 <dbl [45]>## 10 50 -2 <dbl [50]>## # … with 500 more rowsUnnest
conditions %>% as_tibble() %>% mutate(sim = map2(n, mu, ~rnorm(.x, .y, sd = 10))) %>% unnest(sim)## # A tibble: 39,525 × 3## n mu sim## <dbl> <dbl> <dbl>## 1 5 -2 -7.886723## 2 5 -2 5.402129## 3 5 -2 -9.258305## 4 5 -2 -5.295245## 5 5 -2 -17.72833 ## 6 10 -2 13.15974 ## 7 10 -2 -17.04609 ## 8 10 -2 3.520510## 9 10 -2 -18.55369 ## 10 10 -2 5.565162## # … with 39,515 more rowsChallenge
Can you replicate what we just did, but using a rowwise() approach?
03:00
conditions %>% rowwise() %>% mutate(sim = list(rnorm(n, mu, sd = 10))) %>% unnest(sim)## # A tibble: 39,525 × 3## n mu sim## <dbl> <dbl> <dbl>## 1 5 -2 3.591224 ## 2 5 -2 -18.28598 ## 3 5 -2 4.355475 ## 4 5 -2 10.54270 ## 5 5 -2 7.748755 ## 6 10 -2 4.939168 ## 7 10 -2 1.026145 ## 8 10 -2 -13.14307 ## 9 10 -2 0.8444179## 10 10 -2 -8.977694 ## # … with 39,515 more rowsThe data
Please follow along
library(fivethirtyeight)pulitzer## # A tibble: 50 × 7## newspaper circ2004 circ2013 pctchg_circ num_finals1990_2003## <chr> <dbl> <dbl> <int> <int>## 1 USA Today 2192098 1674306 -24 1## 2 Wall Street Journal 2101017 2378827 13 30## 3 New York Times 1119027 1865318 67 55## 4 Los Angeles Times 983727 653868 -34 44## 5 Washington Post 760034 474767 -38 52## 6 New York Daily News 712671 516165 -28 4## 7 New York Post 642844 500521 -22 0## 8 Chicago Tribune 603315 414930 -31 23## 9 San Jose Mercury News 558874 583998 4 4## 10 Newsday 553117 377744 -32 12## # … with 40 more rows, and 2 more variables: num_finals2004_2014 <int>,## # num_finals1990_2014 <int>Prep data
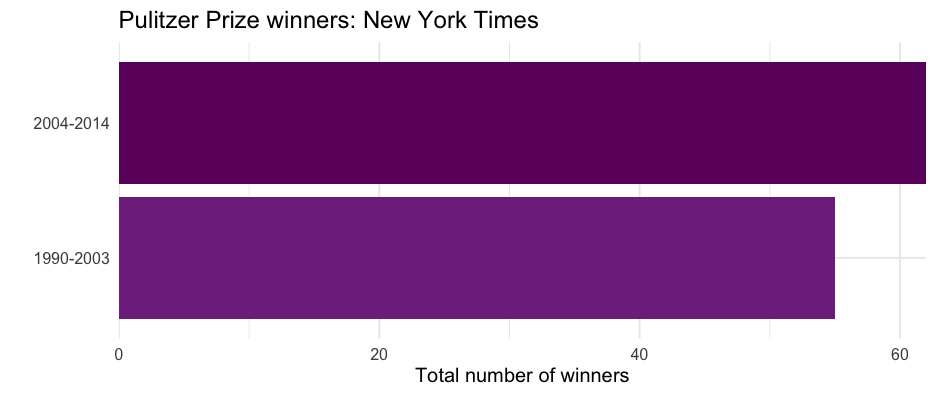
pulitzer<- pulitzer %>% select(newspaper, starts_with("num")) %>% pivot_longer( -newspaper, names_to = "year_range", values_to = "n", names_prefix = "num_finals" ) %>% mutate(year_range = str_replace_all(year_range, "_", "-")) %>% filter(year_range != "1990-2014")head(pulitzer)## # A tibble: 6 × 3## newspaper year_range n## <chr> <chr> <int>## 1 USA Today 1990-2003 1## 2 USA Today 2004-2014 1## 3 Wall Street Journal 1990-2003 30## 4 Wall Street Journal 2004-2014 20## 5 New York Times 1990-2003 55## 6 New York Times 2004-2014 62One plot
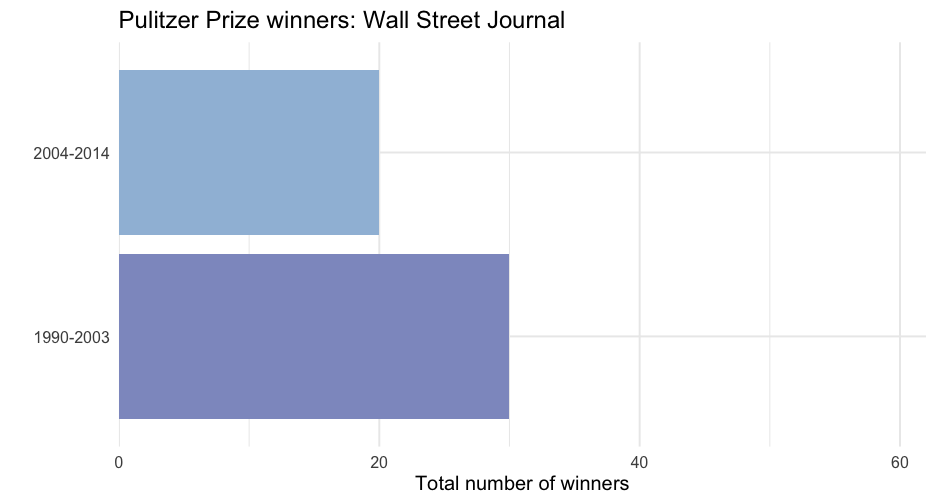
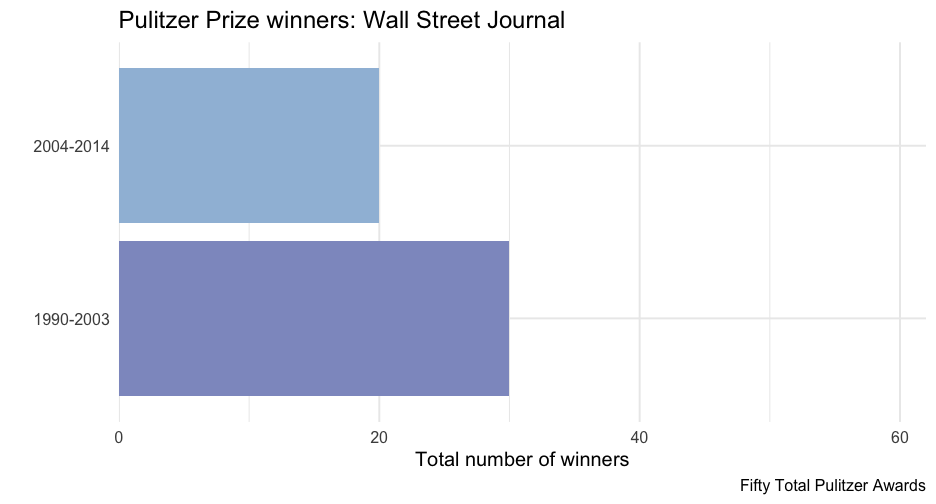
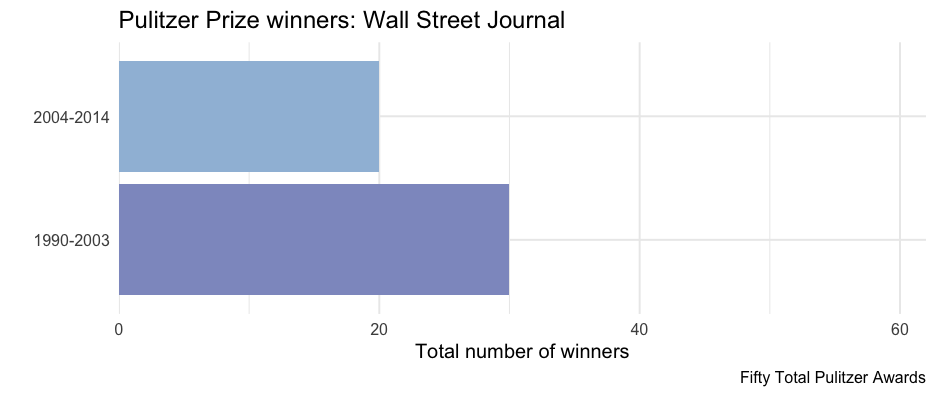
wsj <- pulitzer %>% filter(newspaper == "Wall Street Journal")ggplot(wsj, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = "Pulitzer Prize winners: Wall Street Journal", x = "Total number of winners", y = "" )Nest data
by_newspaper <- pulitzer %>% group_by(newspaper) %>% nest()by_newspaper## # A tibble: 50 × 2## # Groups: newspaper [50]## newspaper data ## <chr> <list> ## 1 USA Today <tibble [2 × 2]>## 2 Wall Street Journal <tibble [2 × 2]>## 3 New York Times <tibble [2 × 2]>## 4 Los Angeles Times <tibble [2 × 2]>## 5 Washington Post <tibble [2 × 2]>## 6 New York Daily News <tibble [2 × 2]>## 7 New York Post <tibble [2 × 2]>## 8 Chicago Tribune <tibble [2 × 2]>## 9 San Jose Mercury News <tibble [2 × 2]>## 10 Newsday <tibble [2 × 2]>## # … with 40 more rowsby_newspaper %>% mutate( plot = map( data, ~{ ggplot(aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = "Pulitzer Prize winners", x = "Total number of winners", y = "" ) } ) )Add title
library(glue)p <- by_newspaper %>% mutate( plot = map2( data, newspaper, ~{ ggplot(.x, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = glue("Pulitzer Prize winners: {.y}"), x = "Total number of winners", y = "" ) } ) )p## # A tibble: 50 × 3## # Groups: newspaper [50]## newspaper data plot ## <chr> <list> <list>## 1 USA Today <tibble [2 × 2]> <gg> ## 2 Wall Street Journal <tibble [2 × 2]> <gg> ## 3 New York Times <tibble [2 × 2]> <gg> ## 4 Los Angeles Times <tibble [2 × 2]> <gg> ## 5 Washington Post <tibble [2 × 2]> <gg> ## 6 New York Daily News <tibble [2 × 2]> <gg> ## 7 New York Post <tibble [2 × 2]> <gg> ## 8 Chicago Tribune <tibble [2 × 2]> <gg> ## 9 San Jose Mercury News <tibble [2 × 2]> <gg> ## 10 Newsday <tibble [2 × 2]> <gg> ## # … with 40 more rowspulitzer %>%nest_by(newspaper) %>% mutate( plot = list( ggplot(data, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = glue("Pulitzer Prize winners: {newspaper}"), x = "Total number of winners", y = "" ) ) )## # A tibble: 50 × 3## # Rowwise: newspaper## newspaper data plot ## <chr> <list<tibble[,2]>> <list>## 1 Arizona Republic [2 × 2] <gg> ## 2 Atlanta Journal Constitution [2 × 2] <gg> ## 3 Baltimore Sun [2 × 2] <gg> ## 4 Boston Globe [2 × 2] <gg> ## 5 Boston Herald [2 × 2] <gg> ## 6 Charlotte Observer [2 × 2] <gg> ## 7 Chicago Sun-Times [2 × 2] <gg> ## 8 Chicago Tribune [2 × 2] <gg> ## 9 Cleveland Plain Dealer [2 × 2] <gg> ## 10 Columbus Dispatch [2 × 2] <gg> ## # … with 40 more rowsfull_conditions <- expand.grid( n = seq(5, 150, 5), mu = seq(-2, 2, 0.25), sd = seq(1, 3, .1))head(full_conditions)## n mu sd## 1 5 -2 1## 2 10 -2 1## 3 15 -2 1## 4 20 -2 1## 5 25 -2 1## 6 30 -2 1tail(full_conditions)## n mu sd## 10705 125 2 3## 10706 130 2 3## 10707 135 2 3## 10708 140 2 3## 10709 145 2 3## 10710 150 2 3Full Simulation
fsim <- pmap( list( number = full_conditions$n, average = full_conditions$mu, stdev = full_conditions$sd ), function(number, average, stdev) { rnorm(n = number, mean = average, sd = stdev) })str(fsim)## List of 10710## $ : num [1:5] -2.899 -1.854 0.271 -0.689 -3.361## $ : num [1:10] -1.043 -1.656 0.428 -2.755 -1.118 ...## $ : num [1:15] -3.508 -2.114 -0.82 -0.952 -1.021 ...## $ : num [1:20] -2.335 -2.774 -3.432 -1.401 -0.857 ...## $ : num [1:25] -2.08 -3.98 -1.8 -1.5 -1.45 ...## $ : num [1:30] -2.51 -2.65 -2.96 -1.85 -2.05 ...## $ : num [1:35] -3.936 -2.175 -0.987 -1.441 -0.568 ...## $ : num [1:40] -1.847 -0.312 0.187 -2.501 -2.88 ...## $ : num [1:45] -3.67 -1.9 -1.29 -3.51 -2.12 ...## $ : num [1:50] -1.693 -2.443 -4.54 -0.818 -1.517 ...## $ : num [1:55] -1.68 -4.4 -3.82 -1.47 -2.61 ...## $ : num [1:60] 0.111 -1.336 -2.077 -1.28 -1.098 ...## $ : num [1:65] -2.57 -2.01 -2.6 -1.93 -3.57 ...## $ : num [1:70] -0.888 -3.818 -1.17 -1.062 -0.615 ...## $ : num [1:75] -1.93 -1.69 -2.49 -1.13 -2.92 ...## $ : num [1:80] -3.464 -1.279 -0.958 -1.72 -1.575 ...## $ : num [1:85] -0.235 -1.901 -2.282 -3.016 -2.692 ...## $ : num [1:90] -1.49 -3.75 -3.51 -2.38 -1.19 ...## $ : num [1:95] -1.92 -2.59 -1.97 -2.54 -1.82 ...## $ : num [1:100] -0.915 -1.657 -2.834 -2.784 -2.876 ...## $ : num [1:105] -1.02 -3.92 -1.86 -1.89 -1.92 ...## $ : num [1:110] -1.7 -1.4 -2.15 -1.03 -1.51 ...## $ : num [1:115] -3.29 -2.86 -0.29 -3.74 -3.79 ...## $ : num [1:120] -3.991 -1.785 -1.635 -0.372 -1.775 ...## $ : num [1:125] -2.1581 -2.0733 -2.6558 0.0774 -2.9995 ...## $ : num [1:130] -2.298 -2.409 -2.41 -1.198 -0.691 ...## $ : num [1:135] -1.762 -2.498 -3.638 0.187 -3.589 ...## $ : num [1:140] -2.79 -2.32 -1.93 -2.14 -2.62 ...## $ : num [1:145] -2.029 -1.438 -0.935 -2.294 -1.775 ...## $ : num [1:150] -2.341 -1.317 -0.486 -1.993 -1.027 ...## $ : num [1:5] -0.439 -1.029 -0.324 0.648 -1.748## $ : num [1:10] -0.81 -1.341 -0.677 -2.591 -1.508 ...## $ : num [1:15] -2.277 -1.561 -0.902 -1.997 -0.446 ...## $ : num [1:20] -1.65 -3.1 -1.45 -2.44 -2.55 ...## $ : num [1:25] -2.957 -0.817 -2.161 -0.57 -1.95 ...## $ : num [1:30] -1.49 -2.08 -1.29 -1.68 -2.38 ...## $ : num [1:35] 0.444 -0.777 -2.673 -0.183 -0.725 ...## $ : num [1:40] -1.081 -2.549 -1.554 -3.196 -0.832 ...## $ : num [1:45] -2.012 -1.392 -2.308 -0.475 -2.15 ...## $ : num [1:50] -1.363 -1.725 -0.641 -0.827 -1.44 ...## $ : num [1:55] -2.497 -0.046 -2.543 -1.318 -2.705 ...## $ : num [1:60] -1.94 -3.78 -3.16 -1.98 -2.56 ...## $ : num [1:65] -2.649 -0.67 -2.373 -1.521 -0.956 ...## $ : num [1:70] -2.454 -1.281 -0.769 -1.251 -2.615 ...## $ : num [1:75] -2.702 -2.445 -1.748 -2.282 -0.503 ...## $ : num [1:80] -3.073 -1.053 -0.248 -1.504 -3.599 ...## $ : num [1:85] -2.389 -1.972 -0.844 -1.913 -0.138 ...## $ : num [1:90] -1.532 -2.292 -1.113 -2.462 -0.612 ...## $ : num [1:95] 0.101 -1.125 -2.077 -2.517 -2.894 ...## $ : num [1:100] -2.501 -1.813 -2.453 -0.454 -2.196 ...## $ : num [1:105] -3.11 -2.16 -3.21 -1.57 -2.29 ...## $ : num [1:110] 0.0221 -0.3542 -1.1348 -1.8837 -2.7147 ...## $ : num [1:115] -2.944 -2.555 -1.606 -3.158 -0.431 ...## $ : num [1:120] -3.05 -1.34 -2.16 -2.17 -2.23 ...## $ : num [1:125] -3.92 -2.43 -2.87 -2.02 -1.96 ...## $ : num [1:130] -1.9806 -1.3754 -0.0422 -1.2931 0.5103 ...## $ : num [1:135] -1.85 -2.443 -0.864 -2.5 -1.318 ...## $ : num [1:140] -3.061 -1.322 -1.618 -0.574 -3.674 ...## $ : num [1:145] -2.95 -1.93 -1.13 -1.3 -2.48 ...## $ : num [1:150] -1.879 -1.544 -0.634 -0.146 -1.209 ...## $ : num [1:5] -0.755 -2.371 -1.138 -0.608 0.11## $ : num [1:10] -1.48 -2.519 -1.476 -1.587 -0.442 ...## $ : num [1:15] -0.621 -1.931 -1.431 0.59 -2.148 ...## $ : num [1:20] -2.474 -1.043 -1.929 -0.483 -0.677 ...## $ : num [1:25] -1.712 -1.314 -2.904 -0.746 -1.364 ...## $ : num [1:30] -1.506 -2.865 -1.779 -2.084 0.218 ...## $ : num [1:35] -0.0888 -2.8476 -1.1306 -1.6184 -1.5593 ...## $ : num [1:40] -0.924 1.252 -0.826 -1.645 -0.794 ...## $ : num [1:45] -1.49 -2.398 -0.421 -1.686 -0.751 ...## $ : num [1:50] -1.051 -0.604 -2.562 -1.946 -0.858 ...## $ : num [1:55] -2.609 0.859 -0.123 -0.503 -2.222 ...## $ : num [1:60] -0.721 -1.289 -1.366 0.449 -1.459 ...## $ : num [1:65] -1.187 -0.731 -0.655 -2.309 -1.116 ...## $ : num [1:70] -1.705 -0.509 -0.645 -0.598 -0.365 ...## $ : num [1:75] -0.165 -1.722 -1.035 -0.533 -1.258 ...## $ : num [1:80] -2.599 0.137 -2.794 -0.989 -1.523 ...## $ : num [1:85] -2.09 -2.47 -1.7 -1.36 -2.08 ...## $ : num [1:90] -0.614 -0.692 -2.174 -2.297 -1.265 ...## $ : num [1:95] -1.27 -0.53 -1.19 -2.7 -1.21 ...## $ : num [1:100] -2.03 -1.4 -3.53 -1.35 -3 ...## $ : num [1:105] -2.239 -0.786 -1.178 -1.509 -1.011 ...## $ : num [1:110] -0.574 -0.255 -3.816 -0.768 -1.251 ...## $ : num [1:115] 0.0849 -0.8844 -2.4786 -2.2744 -1.8509 ...## $ : num [1:120] -0.343 -2.457 -2.097 -3.502 -2.733 ...## $ : num [1:125] -3.766 -1.309 -2.378 -2.602 0.848 ...## $ : num [1:130] -0.759 -3.489 0.05 1.63 -4.049 ...## $ : num [1:135] -3.9458 -0.4551 0.1423 -0.698 -0.0371 ...## $ : num [1:140] -0.971 -2.578 -0.67 -0.472 -2.357 ...## $ : num [1:145] -2.14 -1.8 -2.83 -1.71 -2.34 ...## $ : num [1:150] -0.195 -0.738 -1.85 -0.62 -1.114 ...## $ : num [1:5] -2.424 -1.366 -0.552 -1.418 -1.151## $ : num [1:10] -1.613 -1.826 -0.595 -1.839 0.209 ...## $ : num [1:15] -2.93 -1.74 -1.44 -2.2 -1.38 ...## $ : num [1:20] -1.04 -1.37 -2.55 -1.79 -2.94 ...## $ : num [1:25] -0.7343 -1.7964 0.2689 -0.6538 0.0879 ...## $ : num [1:30] -0.997 -0.948 -1.613 -1.011 -1.937 ...## $ : num [1:35] -3.033 -0.224 -2.761 -1.277 -1.088 ...## $ : num [1:40] -1.118 0.115 -0.618 -0.619 -1.53 ...## $ : num [1:45] -0.318 -0.992 -0.196 -2.175 -0.504 ...## [list output truncated]Alternative spec
fsim <- pmap( list( full_conditions$n, full_conditions$mu, full_conditions$sd ), ~rnorm(n = ..1, mean = ..2, sd = ..3))str(fsim)## List of 10710## $ : num [1:5] -3.392 -0.9933 -2.2558 0.0526 -3.939## $ : num [1:10] -3.24 -2.35 -2.15 -2.74 -2.23 ...## $ : num [1:15] -1.926 -2.1026 -0.0394 -1.9635 -1.8651 ...## $ : num [1:20] -3.177 -2.868 -2.569 -0.733 -0.395 ...## $ : num [1:25] -0.988 -1.228 -0.716 -2.742 -1.866 ...## $ : num [1:30] -3.62 -1.87 -3.83 -1.87 -3.8 ...## $ : num [1:35] -1.623 -1.417 -1.914 -4.289 -0.918 ...## $ : num [1:40] -1.278 -1.693 -1.257 -0.321 -1.72 ...## $ : num [1:45] 0.7488 0.0512 -1.9767 -2.7075 -2.1823 ...## $ : num [1:50] -2.21 -1.88 -2.45 -3.29 -2.66 ...## $ : num [1:55] -3.786 -1.132 -1.259 -0.535 -1.946 ...## $ : num [1:60] -2.972 -2.073 -2.069 -0.997 -3.302 ...## $ : num [1:65] -1.38 -1.56 -3.17 -1.45 -4.86 ...## $ : num [1:70] -3.05 -2.26 -1.57 -2.77 -1.95 ...## $ : num [1:75] -1.713 -3.604 -2.198 -0.497 -2.063 ...## $ : num [1:80] -3.66 -1.33 -2.38 -2.23 -1.59 ...## $ : num [1:85] -1.806 -0.85 -0.685 -2.287 -2.127 ...## $ : num [1:90] -1.952 -3.123 -1.189 0.465 -2.299 ...## $ : num [1:95] -3.05 -1.35 -2.09 -2.65 -1.94 ...## $ : num [1:100] -1.592 -1.421 -1.796 -0.967 -1.348 ...## $ : num [1:105] -0.618 -2.875 -3.457 -3.695 -3.547 ...## $ : num [1:110] -1.3 -2.01 -2.9 -2.54 -4.08 ...## $ : num [1:115] -3.65 -1.27 -2.19 -2.18 -4.18 ...## $ : num [1:120] -3.6812 -1.4201 -0.7788 0.0428 -1.3453 ...## $ : num [1:125] -3.361 -0.919 -1.744 -1.21 -0.925 ...## $ : num [1:130] -1.32 -1.04 -2.61 -3.2 -2.57 ...## $ : num [1:135] -2.5 -1.49 -3.16 -1.11 -1.96 ...## $ : num [1:140] -2.21 -2.7 -2.91 -1.77 -1.03 ...## $ : num [1:145] -2.524 -0.335 -2.964 -2.305 -2.409 ...## $ : num [1:150] -1.977 -0.296 -0.799 -2.052 -1.649 ...## $ : num [1:5] -1.44 -1.79 -1.2 -2.14 -2.21## $ : num [1:10] -1.208 -1.917 -0.693 -2.271 -3.184 ...## $ : num [1:15] -2.794 -2.852 -0.838 -2.227 0.829 ...## $ : num [1:20] -3.094 -1.653 -0.407 -1.516 -1.854 ...## $ : num [1:25] -3.1792 0.0503 -1.46 -2.3736 -1.208 ...## $ : num [1:30] -0.345 -3.195 -1.637 -0.839 -1.329 ...## $ : num [1:35] -1.63 -1.95 -1.18 -3.84 -2.96 ...## $ : num [1:40] -1.48 0.11 -0.252 -1.887 0.639 ...## $ : num [1:45] -2.022 -1.631 -0.956 -0.545 -2.216 ...## $ : num [1:50] -2.129 -1.019 -0.248 -2.113 -2.491 ...## $ : num [1:55] -2.5323 0.0926 -1.103 -0.7597 -1.282 ...## $ : num [1:60] -2.25 -1.92 -1.01 -1.9 -2.89 ...## $ : num [1:65] -1.28 -0.361 -3.539 -1.221 -1.969 ...## $ : num [1:70] -1.772 -0.605 -2.142 -0.841 -2.011 ...## $ : num [1:75] -1.671 -1.638 -1.456 -0.584 -1.285 ...## $ : num [1:80] -0.519 -3.402 -1.545 -1.815 -1.021 ...## $ : num [1:85] -1.861 -2.997 0.904 -3.716 -2.798 ...## $ : num [1:90] -2.86 -1.54 -1.49 -1.37 -2.32 ...## $ : num [1:95] -2.91 -1.85 -4.96 -2.08 -2.47 ...## $ : num [1:100] -2.24 -0.384 -2.815 -1.314 -0.898 ...## $ : num [1:105] -1.5 -1.52 -1.79 -1.23 -2.85 ...## $ : num [1:110] -0.199 -1.538 -2.178 -2.888 -0.99 ...## $ : num [1:115] -0.428 -1.553 -2.157 -2.47 -2.429 ...## $ : num [1:120] 1.445 -1.915 0.103 -2.584 -2.88 ...## $ : num [1:125] -1.756 -2.689 -2.043 -1.676 -0.454 ...## $ : num [1:130] -1.68 -2.38 -1.46 -2.39 -1.42 ...## $ : num [1:135] -1.946 -0.747 -0.493 -2.931 -1.555 ...## $ : num [1:140] -0.159 -2.58 -2.861 -2.188 -0.117 ...## $ : num [1:145] -1.31 -1.92 -1.56 -1.36 -2.28 ...## $ : num [1:150] -1.19 -2.03 -3.39 -2.76 -2.17 ...## $ : num [1:5] -1.755 -0.144 -1.142 -1.182 -0.973## $ : num [1:10] -0.058 -1.808 -1.246 -0.48 -1.446 ...## $ : num [1:15] -3.24 -1.07 -3.14 -2.23 -2.05 ...## $ : num [1:20] -2.285 -1.284 -2.366 -2.14 -0.777 ...## $ : num [1:25] -2.7 -2.42 -3.54 -2.58 -1.68 ...## $ : num [1:30] -1.331 -1.573 -1.796 -0.855 -2.431 ...## $ : num [1:35] -0.94 -1.457 -0.947 -3.221 -0.638 ...## $ : num [1:40] -1.842 -2.925 -0.187 -1.049 -1.066 ...## $ : num [1:45] 1.073 -3.284 -0.997 -1.295 -2.392 ...## $ : num [1:50] -2.934 0.483 -0.458 0.616 -0.465 ...## $ : num [1:55] -1.26 -2.682 -1.536 -0.923 -0.653 ...## $ : num [1:60] -2.905 -1.504 -3.405 -0.807 -1.185 ...## $ : num [1:65] -2.626 -2.5 -1.837 -0.205 -3.061 ...## $ : num [1:70] -2.541 0.113 -1.637 -1.397 -0.749 ...## $ : num [1:75] -1.15 -1.38 -2.01 -2.28 -1.6 ...## $ : num [1:80] -0.271 -2.957 -1.095 -0.255 0.186 ...## $ : num [1:85] -2.169 -1.762 -2.371 -2.084 -0.762 ...## $ : num [1:90] -1.2019 -3.7622 -2.6474 -0.0599 -2.1553 ...## $ : num [1:95] -1.316 -1.797 -1.518 -0.539 0.496 ...## $ : num [1:100] -2.65 -2 -1.31 -2.24 -2.1 ...## $ : num [1:105] -3.51 -1.9 -1.42 -3.59 -3.35 ...## $ : num [1:110] -2.267 -2.803 -0.447 -2.074 -2.612 ...## $ : num [1:115] -2.04 -1.89 -1.85 -1.22 -3.31 ...## $ : num [1:120] -1.55 -0.677 -1.538 1.082 -1.294 ...## $ : num [1:125] -0.1396 -0.5911 -1.481 -2.5253 -0.0471 ...## $ : num [1:130] -0.951 -0.901 -0.713 -1.634 -0.859 ...## $ : num [1:135] -2.73 -3.72 -2.23 -2.36 -2.11 ...## $ : num [1:140] -2.487 -1.0681 -0.0279 -1.9474 -0.6085 ...## $ : num [1:145] -0.231 -2.073 -1.83 -2.429 -1.515 ...## $ : num [1:150] 0.457 -1.126 0.306 -2.274 -1.24 ...## $ : num [1:5] -0.331 -1.507 -3.153 -0.757 -3.085## $ : num [1:10] -1.451 -2.849 -1.588 -0.859 0.993 ...## $ : num [1:15] -2.511 -1.289 -2.724 -0.506 0.896 ...## $ : num [1:20] -3.386 -0.231 -2.41 -4.034 -1.106 ...## $ : num [1:25] -2.16 -0.614 0.462 -1.392 -1.012 ...## $ : num [1:30] -1.389 -0.523 -0.267 -0.41 -2.295 ...## $ : num [1:35] -0.509 -1.428 -1.303 -1.684 -1.458 ...## $ : num [1:40] -1.561 0.118 -2.219 -1.718 -0.434 ...## $ : num [1:45] -0.606 -1.276 -3.395 -1.145 -2.504 ...## [list output truncated]Simpler
Maybe a little too clever
- A data frame is a list so...
fsim <- pmap( full_conditions, ~rnorm(n = ..1, mean = ..2, sd = ..3))str(fsim)## List of 10710## $ : num [1:5] -2.483 -0.966 -1.195 -3.669 -0.783## $ : num [1:10] -3.01 -1.753 -2.453 -0.996 -1.681 ...## $ : num [1:15] -3.51 -3.02 -2.73 -3.52 -2.46 ...## $ : num [1:20] -3.01 -2.1 -2.31 -1.5 -0.67 ...## $ : num [1:25] -1.418 -3.201 -1.511 0.791 -3.146 ...## $ : num [1:30] -0.652 -1.063 -2.132 -1.504 -1.339 ...## $ : num [1:35] -3.207 -1.448 -2.173 -2.473 -0.681 ...## $ : num [1:40] -3.41 -1.69 -4.06 -3.56 -2.22 ...## $ : num [1:45] -3.17 -1.91 -1.43 -3.02 -2.35 ...## $ : num [1:50] -2.65 -3.61 -2.39 -1.36 -1.04 ...## $ : num [1:55] -2.62 -1.63 -2.51 -2 -2.6 ...## $ : num [1:60] -3.83 -2.84 -2.93 -1.56 -2.7 ...## $ : num [1:65] -1.0858 -3 -2.0788 0.0333 -2.8598 ...## $ : num [1:70] -1.91 -2.29 -2.09 -2.36 -2.76 ...## $ : num [1:75] -0.903 -4.198 -1.353 -1.853 -3.146 ...## $ : num [1:80] -2.82 -1.66 -2.23 -2.56 -1.73 ...## $ : num [1:85] -1.38 -2.484 -2.869 -3.29 -0.604 ...## $ : num [1:90] -1.34 -2.65 -2 -1.45 -1.61 ...## $ : num [1:95] -2.15 -1.31 -1.58 -2.11 -2.06 ...## $ : num [1:100] -3.367 -0.512 -2.598 -2.212 -4.086 ...## $ : num [1:105] -0.144 -0.531 -2.808 -1.923 -2.798 ...## $ : num [1:110] -1.92 -3.04 -3.57 -1.04 -3.64 ...## $ : num [1:115] -1.959 -0.664 -2.037 -2.64 -3.506 ...## $ : num [1:120] -1.4242 -3.6394 0.0901 -0.9878 -3.4518 ...## $ : num [1:125] -0.4124 0.0652 -2.0728 -2.5879 -1.2572 ...## $ : num [1:130] -1.73 -1.1 -2.54 -1.75 -2.43 ...## $ : num [1:135] -3.037 0.758 -3.231 -1.685 -2.654 ...## $ : num [1:140] -2.51 -1.38 -2.37 -3.38 -3.79 ...## $ : num [1:145] -2.04 -1.93 -2.6 -3.45 -2.38 ...## $ : num [1:150] -1.89 -1.67 -2.82 -3.75 -1.16 ...## $ : num [1:5] -2.123 -1.03 -0.132 -2.666 -2.68## $ : num [1:10] -2.37 -3.44 -1.01 -2.5 -1.65 ...## $ : num [1:15] -0.644 -2.291 -1.436 -1.098 -1.4 ...## $ : num [1:20] -1.38 -2.38 -1.21 -1.94 -2.12 ...## $ : num [1:25] -2.7613 -2.0795 -0.3011 -0.0335 -2.3788 ...## $ : num [1:30] -0.302 -2.515 -1.307 -2.251 -2.374 ...## $ : num [1:35] -1.255 -3.153 -0.839 -1.495 -2.032 ...## $ : num [1:40] -0.601 -1.763 -1.842 -0.172 -0.766 ...## $ : num [1:45] -2.293 -0.181 0.824 -1.397 -3.079 ...## $ : num [1:50] -2.94 -2.37 -2.18 -1.56 -2.2 ...## $ : num [1:55] -2.232 -0.997 -1.977 -2.326 -2.458 ...## $ : num [1:60] -1.055 -2.254 -0.833 -1.18 -1.043 ...## $ : num [1:65] -2.96 -1.5 -2.46 -2.14 -2.84 ...## $ : num [1:70] 0.94 -0.713 -2.682 -2.092 -1.614 ...## $ : num [1:75] -1.806 -1.756 -3.262 -0.767 -1.556 ...## $ : num [1:80] -1.495 -2.852 -1.99 -0.24 -0.343 ...## $ : num [1:85] -1.75 -0.965 -0.194 -0.576 -1.734 ...## $ : num [1:90] -1.71 -1.13 -1.85 -2.72 -1.81 ...## $ : num [1:95] -2.432 -1.533 -2.132 -4.217 -0.904 ...## $ : num [1:100] -1.716 -0.384 -1.252 -2.841 -2.544 ...## $ : num [1:105] -1.11 -2.88 -1.67 -2.48 -1.29 ...## $ : num [1:110] 0.815 -2.386 0.376 -1.338 -2.056 ...## $ : num [1:115] -0.937 -0.926 -2.475 -1.613 -1.559 ...## $ : num [1:120] -0.111 -1.958 -3.896 -1.69 -0.475 ...## $ : num [1:125] -2.072 -2.736 -1.115 -2.031 -0.787 ...## $ : num [1:130] 0.0353 -1.6196 -3.5122 -1.7421 -1.1754 ...## $ : num [1:135] -1.577 -1.956 -0.128 -0.582 -1.9 ...## $ : num [1:140] -2.278 -1.32 -1.607 -1.771 -0.969 ...## $ : num [1:145] -1.0544 -2.6704 -2.4971 0.0864 -1.2751 ...## $ : num [1:150] -2.187 0.888 0.393 -2.528 -2.041 ...## $ : num [1:5] -1.272 -0.69 -0.406 -2.852 -1.571## $ : num [1:10] -0.751 -3.165 -1.587 -1.577 -1.436 ...## $ : num [1:15] -2.21 -1.18 -1.08 -1.1 -2.09 ...## $ : num [1:20] -2.604 -1.13 -1.189 -0.248 -1.348 ...## $ : num [1:25] -2.191 -1.821 -0.81 -0.224 -1.624 ...## $ : num [1:30] -1.686 -0.974 -2.65 -0.6 -0.3 ...## $ : num [1:35] -2.69 -2.763 -1.149 -0.568 -1.606 ...## $ : num [1:40] -1.97 -1.82 -2.35 -1.36 -2.66 ...## $ : num [1:45] -1.05 -1.25 -2.49 -2.08 -2.02 ...## $ : num [1:50] -1.662 -2.085 -2.097 -2.392 0.679 ...## $ : num [1:55] -0.299 -1.191 -1.685 -2.122 -1.134 ...## $ : num [1:60] -0.855 -1.275 -1.451 -2.034 -2.878 ...## $ : num [1:65] -0.239 0.171 -1.282 -1.097 0.428 ...## $ : num [1:70] -1.956 -0.806 -1.187 -1.751 -3.6 ...## $ : num [1:75] 0.12 -2.59 -2.3 -1.28 -1.65 ...## $ : num [1:80] -0.986 -1.648 -0.997 -1.535 -0.459 ...## $ : num [1:85] -1.583 -1.975 -0.728 -1.058 -1.773 ...## $ : num [1:90] -0.0379 -0.5056 -2.181 -2.4305 -1.6892 ...## $ : num [1:95] -1.68 -2.994 -1.427 -0.961 -1.526 ...## $ : num [1:100] -1.05 -2.78 -0.82 -2.84 -1.04 ...## $ : num [1:105] -2.075 -2.64 -0.456 -2.199 -1.442 ...## $ : num [1:110] -1.93 -1.75 -2.65 -2.03 -2.44 ...## $ : num [1:115] -0.511 -0.668 -0.715 -0.494 -0.854 ...## $ : num [1:120] -1.248 -2.938 -0.263 -0.434 -2.303 ...## $ : num [1:125] -1.454 -1.68 0.159 -1.013 -1.026 ...## $ : num [1:130] -0.109 -0.339 -0.806 -0.189 -1.976 ...## $ : num [1:135] -0.766 -2.496 -1.339 -0.876 -1.522 ...## $ : num [1:140] -2.132 -3.443 -1.559 -2.438 -0.533 ...## $ : num [1:145] -0.305 -2.488 0.503 -2.006 -1.59 ...## $ : num [1:150] -1.906 -3.717 -2.481 -0.332 -1.673 ...## $ : num [1:5] 0.0907 -2.4147 -0.2001 -0.3707 -3.2347## $ : num [1:10] -2.37 -0.236 -0.542 -1.363 -1.766 ...## $ : num [1:15] -1.275 -1.384 -0.375 -1.371 -3.41 ...## $ : num [1:20] -1.077 -1.299 -1.341 -0.526 -2.133 ...## $ : num [1:25] -0.893 -2.269 -2.76 -1.079 -2.815 ...## $ : num [1:30] -1.252 -2.188 -0.463 -0.432 -3.33 ...## $ : num [1:35] -1.94 -1.86 -1.07 -2.79 -0.39 ...## $ : num [1:40] -0.698 -0.343 -1.36 -0.426 -0.569 ...## $ : num [1:45] -0.282 -0.528 -1.024 -1.173 -2.988 ...## [list output truncated]List column version
full_conditions %>% as_tibble() %>% mutate(sim = pmap(list(n, mu, sd), ~rnorm(..1, ..2, ..3)))## # A tibble: 10,710 × 4## n mu sd sim ## <dbl> <dbl> <dbl> <list> ## 1 5 -2 1 <dbl [5]> ## 2 10 -2 1 <dbl [10]>## 3 15 -2 1 <dbl [15]>## 4 20 -2 1 <dbl [20]>## 5 25 -2 1 <dbl [25]>## 6 30 -2 1 <dbl [30]>## 7 35 -2 1 <dbl [35]>## 8 40 -2 1 <dbl [40]>## 9 45 -2 1 <dbl [45]>## 10 50 -2 1 <dbl [50]>## # … with 10,700 more rowsUnnest
full_conditions %>% as_tibble() %>% mutate(sim = pmap( list(n, mu, sd), ~rnorm(..1, ..2, ..3) ) ) %>% unnest(sim)## # A tibble: 830,025 × 4## n mu sd sim## <dbl> <dbl> <dbl> <dbl>## 1 5 -2 1 -1.496691 ## 2 5 -2 1 -3.955064 ## 3 5 -2 1 -2.074510 ## 4 5 -2 1 -3.654000 ## 5 5 -2 1 -2.049551 ## 6 10 -2 1 -2.332229 ## 7 10 -2 1 -3.707039 ## 8 10 -2 1 -2.192668 ## 9 10 -2 1 -2.232620 ## 10 10 -2 1 -0.4610839## # … with 830,015 more rowsReplicate with nest_by()
You try first
03:00
full_conditions %>% rowwise() %>% mutate(sim = list(rnorm(n, mu, sd))) %>% unnest(sim)## # A tibble: 830,025 × 4## n mu sd sim## <dbl> <dbl> <dbl> <dbl>## 1 5 -2 1 -0.8569708## 2 5 -2 1 -2.321645 ## 3 5 -2 1 -1.759086 ## 4 5 -2 1 -2.139428 ## 5 5 -2 1 -2.602792 ## 6 10 -2 1 -0.9650236## 7 10 -2 1 -2.013695 ## 8 10 -2 1 -2.970106 ## 9 10 -2 1 -2.655783 ## 10 10 -2 1 -1.272286 ## # … with 830,015 more rowsAdd column for total
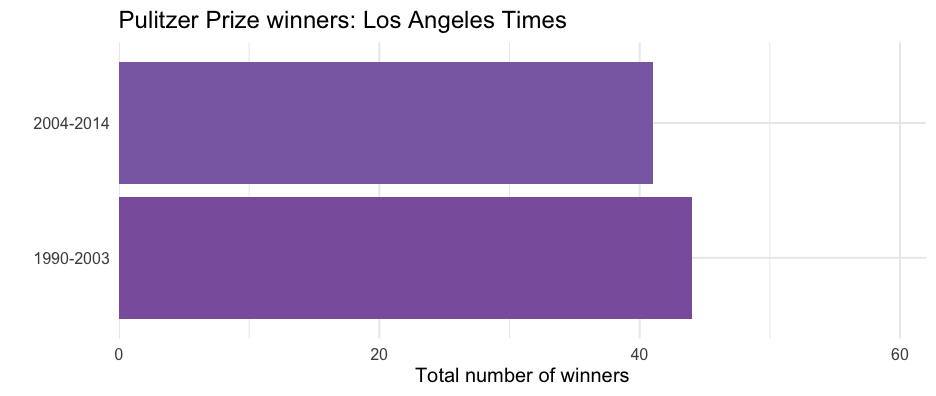
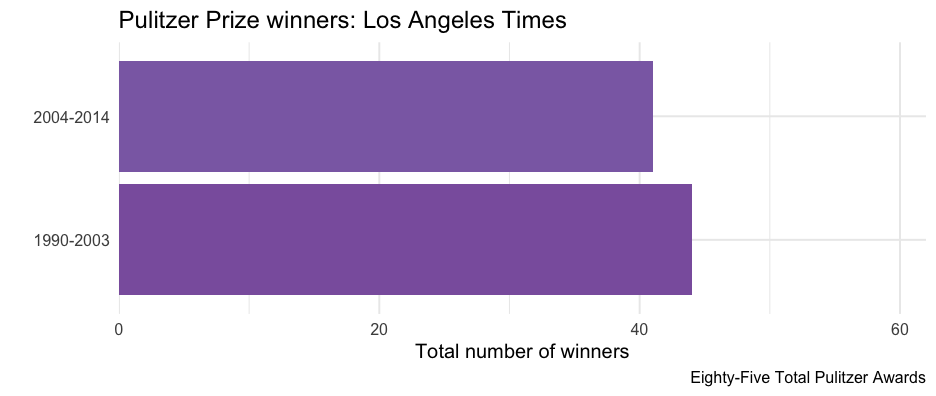
pulitzer <- pulitzer %>% group_by(newspaper) %>% mutate(tot = sum(n))pulitzer## # A tibble: 100 × 4## # Groups: newspaper [50]## newspaper year_range n tot## <chr> <chr> <int> <int>## 1 USA Today 1990-2003 1 2## 2 USA Today 2004-2014 1 2## 3 Wall Street Journal 1990-2003 30 50## 4 Wall Street Journal 2004-2014 20 50## 5 New York Times 1990-2003 55 117## 6 New York Times 2004-2014 62 117## 7 Los Angeles Times 1990-2003 44 85## 8 Los Angeles Times 2004-2014 41 85## 9 Washington Post 1990-2003 52 100## 10 Washington Post 2004-2014 48 100## # … with 90 more rowsselect(pulitzer, newspaper, label)## # A tibble: 100 × 2## # Groups: newspaper [50]## newspaper label ## <chr> <glue> ## 1 USA Today Two Total Pulitzer Awards ## 2 USA Today Two Total Pulitzer Awards ## 3 Wall Street Journal Fifty Total Pulitzer Awards ## 4 Wall Street Journal Fifty Total Pulitzer Awards ## 5 New York Times One Hundred Seventeen Total Pulitzer Awards## 6 New York Times One Hundred Seventeen Total Pulitzer Awards## 7 Los Angeles Times Eighty-Five Total Pulitzer Awards ## 8 Los Angeles Times Eighty-Five Total Pulitzer Awards ## 9 Washington Post One Hundred Total Pulitzer Awards ## 10 Washington Post One Hundred Total Pulitzer Awards ## # … with 90 more rowsProduce one plot
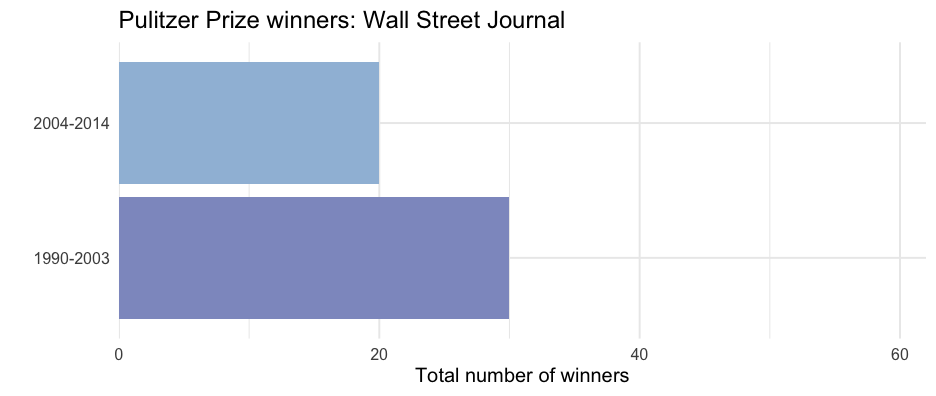
wsj2 <- pulitzer %>% filter(newspaper == "Wall Street Journal") ggplot(wsj2, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = glue("Pulitzer Prize winners: Wall Street Journal"), x = "Total number of winners", y = "", caption = unique(wsj2$label) )Produce all plots
Nest first
by_newspaper_label <- pulitzer %>% group_by(newspaper, label) %>% nest() by_newspaper_label## # A tibble: 50 × 3## # Groups: newspaper, label [50]## newspaper label data ## <chr> <glue> <list> ## 1 USA Today Two Total Pulitzer Awards <tibble [2 × 3]>## 2 Wall Street Journal Fifty Total Pulitzer Awards <tibble [2 × 3]>## 3 New York Times One Hundred Seventeen Total Pulitzer Awards <tibble [2 × 3]>## 4 Los Angeles Times Eighty-Five Total Pulitzer Awards <tibble [2 × 3]>## 5 Washington Post One Hundred Total Pulitzer Awards <tibble [2 × 3]>## 6 New York Daily News Six Total Pulitzer Awards <tibble [2 × 3]>## 7 New York Post Zero Total Pulitzer Awards <tibble [2 × 3]>## 8 Chicago Tribune Thirty-Eight Total Pulitzer Awards <tibble [2 × 3]>## 9 San Jose Mercury News Six Total Pulitzer Awards <tibble [2 × 3]>## 10 Newsday Eighteen Total Pulitzer Awards <tibble [2 × 3]>## # … with 40 more rowsProduce plots
final_plots <- by_newspaper_label %>% mutate(plots = pmap(list(newspaper, label, data), ~{ ggplot(..3, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = glue("Pulitzer Prize winners: {..1}"), x = "Total number of winners", y = "", caption = ..2 ) }))final_plots2 <- pulitzer %>% ungroup() %>% nest_by(newspaper, label) %>% mutate( plots = list( ggplot(data, aes(n, year_range)) + geom_col(aes(fill = n)) + scale_fill_distiller( type = "seq", limits = c(0, max(pulitzer$n)), palette = "BuPu", direction = 1 ) + scale_x_continuous( limits = c(0, max(pulitzer$n)), expand = c(0, 0) ) + guides(fill = "none") + labs( title = glue("Pulitzer Prize winners: {newspaper}"), x = "Total number of winners", y = "", caption = label ) ))final_plots2## # A tibble: 50 × 4## # Rowwise: newspaper, label## newspaper label data plots## <chr> <glue> <list<tibble[> <lis>## 1 Arizona Republic Seven Total Pulitzer Awards [2 × 3] <gg> ## 2 Atlanta Journal Constitution Six Total Pulitzer Awards [2 × 3] <gg> ## 3 Baltimore Sun Thirteen Total Pulitzer Awards [2 × 3] <gg> ## 4 Boston Globe Forty-One Total Pulitzer Awards [2 × 3] <gg> ## 5 Boston Herald Zero Total Pulitzer Awards [2 × 3] <gg> ## 6 Charlotte Observer Four Total Pulitzer Awards [2 × 3] <gg> ## 7 Chicago Sun-Times Two Total Pulitzer Awards [2 × 3] <gg> ## 8 Chicago Tribune Thirty-Eight Total Pulitzer Awards [2 × 3] <gg> ## 9 Cleveland Plain Dealer Eleven Total Pulitzer Awards [2 × 3] <gg> ## 10 Columbus Dispatch One Total Pulitzer Awards [2 × 3] <gg> ## # … with 40 more rowsSave all plots
We'll have to iterate across at least two things: (a) file path/names, and (b) the plots themselves
We can do this with the map() family, but instead we'll use a different function, which we'll talk about more momentarily.
As an aside, what are the steps we would need to take to do this?
Save all plots
We'll have to iterate across at least two things: (a) file path/names, and (b) the plots themselves
We can do this with the map() family, but instead we'll use a different function, which we'll talk about more momentarily.
As an aside, what are the steps we would need to take to do this?
Could we use a nest_by() solution?
Example
Create a directory
fs::dir_create(here::here("plots", "pulitzers"))Create file paths
files <- str_replace_all( tolower(final_plots$newspaper), " ", "-")paths <- here::here("plots", "pulitzers", glue("{files}.png"))paths## [1] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/usa-today.png" ## [2] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/wall-street-journal.png" ## [3] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/new-york-times.png" ## [4] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/los-angeles-times.png" ## [5] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/washington-post.png" ## [6] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/new-york-daily-news.png" ## [7] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/new-york-post.png" ## [8] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/chicago-tribune.png" ## [9] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/san-jose-mercury-news.png" ## [10] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/newsday.png" ## [11] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/houston-chronicle.png" ## [12] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/dallas-morning-news.png" ## [13] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/san-francisco-chronicle.png" ## [14] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/arizona-republic.png" ## [15] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/chicago-sun-times.png" ## [16] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/boston-globe.png" ## [17] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/atlanta-journal-constitution.png"## [18] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/newark-star-ledger.png" ## [19] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/detroit-free-press.png" ## [20] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/minneapolis-star-tribune.png" ## [21] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/philadelphia-inquirer.png" ## [22] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/cleveland-plain-dealer.png" ## [23] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/san-diego-union-tribune.png" ## [24] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/tampa-bay-times.png" ## [25] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/denver-post.png" ## [26] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/rocky-mountain-news.png" ## [27] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/oregonian.png" ## [28] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/miami-herald.png" ## [29] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/orange-county-register.png" ## [30] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/sacramento-bee.png" ## [31] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/st.-louis-post-dispatch.png" ## [32] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/baltimore-sun.png" ## [33] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/kansas-city-star.png" ## [34] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/detroit-news.png" ## [35] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/orlando-sentinel.png" ## [36] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/south-florida-sun-sentinel.png" ## [37] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/new-orleans-times-picayune.png" ## [38] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/columbus-dispatch.png" ## [39] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/indianapolis-star.png" ## [40] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/san-antonio-express-news.png" ## [41] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/pittsburgh-post-gazette.png" ## [42] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/milwaukee-journal-sentinel.png" ## [43] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/tampa-tribune.png" ## [44] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/fort-woth-star-telegram.png" ## [45] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/boston-herald.png" ## [46] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/seattle-times.png" ## [47] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/charlotte-observer.png" ## [48] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/daily-oklahoman.png" ## [49] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/louisville-courier-journal.png" ## [50] "/Users/daniel/Teaching/data_sci_specialization/2021-22/c3-fp-2022/plots/pulitzers/investor's-buisiness-daily.png"Add paths to data frame
final_plots %>% ungroup() %>% mutate(path = paths) %>% select(plots, path)## # A tibble: 50 × 2## plots ## <list>## 1 <gg> ## 2 <gg> ## 3 <gg> ## 4 <gg> ## 5 <gg> ## 6 <gg> ## 7 <gg> ## 8 <gg> ## 9 <gg> ## 10 <gg> ## # … with 40 more rows, and 1 more variable: path <chr>Wrap-up
Parallel iterations greatly increase the things you can do - iterating through at least two things simultaneously is pretty common
The
nest_by()approach can regularly get you the same result asgroup_by() %>% nest() %>% mutate() %>% map()- Caveat - must be in a data frame, which means working with list columns
Wrap-up
Parallel iterations greatly increase the things you can do - iterating through at least two things simultaneously is pretty common
The
nest_by()approach can regularly get you the same result asgroup_by() %>% nest() %>% mutate() %>% map()- Caveat - must be in a data frame, which means working with list columns
- My view - it's still worth learning both. Looping with {purrr} is super flexible and often safer than base versions (type safe). Doesn't have to be used within a data frame.
Reminder
Learning Objectives (for this part)
Know when to apply
walkinstead ofmap, and why it may be usefulUnderstand the parallels and differences between
mapandmodifyDiagnose errors with
safelyand understand other situations where it may be helpfulCollapsing/reducing lists with
purrr::reduce()orbase::Reduce()
Why walk()?
Walk is an alternative to map that you use when you want to call a function for its side effects, rather than for its return value. You typically do this because you want to render output to the screen or save files to disk - the important thing is the action, not the return value.
-r4ds
map vs modify
map
map(mtcars, ~as.numeric(scale(.x)))## $mpg## [1] 0.15088482 0.15088482 0.44954345 0.21725341 -0.23073453 -0.33028740## [7] -0.96078893 0.71501778 0.44954345 -0.14777380 -0.38006384 -0.61235388## [13] -0.46302456 -0.81145962 -1.60788262 -1.60788262 -0.89442035 2.04238943## [19] 1.71054652 2.29127162 0.23384555 -0.76168319 -0.81145962 -1.12671039## [25] -0.14777380 1.19619000 0.98049211 1.71054652 -0.71190675 -0.06481307## [31] -0.84464392 0.21725341## ## $cyl## [1] -0.1049878 -0.1049878 -1.2248578 -0.1049878 1.0148821 -0.1049878 1.0148821## [8] -1.2248578 -1.2248578 -0.1049878 -0.1049878 1.0148821 1.0148821 1.0148821## [15] 1.0148821 1.0148821 1.0148821 -1.2248578 -1.2248578 -1.2248578 -1.2248578## [22] 1.0148821 1.0148821 1.0148821 1.0148821 -1.2248578 -1.2248578 -1.2248578## [29] 1.0148821 -0.1049878 1.0148821 -1.2248578## ## $disp## [1] -0.57061982 -0.57061982 -0.99018209 0.22009369 1.04308123 -0.04616698## [7] 1.04308123 -0.67793094 -0.72553512 -0.50929918 -0.50929918 0.36371309## [13] 0.36371309 0.36371309 1.94675381 1.84993175 1.68856165 -1.22658929## [19] -1.25079481 -1.28790993 -0.89255318 0.70420401 0.59124494 0.96239618## [25] 1.36582144 -1.22416874 -0.89093948 -1.09426581 0.97046468 -0.69164740## [31] 0.56703942 -0.88529152## ## $hp## [1] -0.53509284 -0.53509284 -0.78304046 -0.53509284 0.41294217 -0.60801861## [7] 1.43390296 -1.23518023 -0.75387015 -0.34548584 -0.34548584 0.48586794## [13] 0.48586794 0.48586794 0.85049680 0.99634834 1.21512565 -1.17683962## [19] -1.38103178 -1.19142477 -0.72469984 0.04831332 0.04831332 1.43390296## [25] 0.41294217 -1.17683962 -0.81221077 -0.49133738 1.71102089 0.41294217## [31] 2.74656682 -0.54967799## ## $drat## [1] 0.56751369 0.56751369 0.47399959 -0.96611753 -0.83519779 -1.56460776## [7] -0.72298087 0.17475447 0.60491932 0.60491932 0.60491932 -0.98482035## [13] -0.98482035 -0.98482035 -1.24665983 -1.11574009 -0.68557523 0.90416444## [19] 2.49390411 1.16600392 0.19345729 -1.56460776 -0.83519779 0.24956575## [25] -0.96611753 0.90416444 1.55876313 0.32437703 1.16600392 0.04383473## [31] -0.10578782 0.96027290## ## $wt## [1] -0.610399567 -0.349785269 -0.917004624 -0.002299538 0.227654255 0.248094592## [7] 0.360516446 -0.027849959 -0.068730634 0.227654255 0.227654255 0.871524874## [13] 0.524039143 0.575139986 2.077504765 2.255335698 2.174596366 -1.039646647## [19] -1.637526508 -1.412682800 -0.768812180 0.309415603 0.222544170 0.636460997## [25] 0.641571082 -1.310481114 -1.100967659 -1.741772228 -0.048290296 -0.457097039## [31] 0.360516446 -0.446876870## ## $qsec## [1] -0.77716515 -0.46378082 0.42600682 0.89048716 -0.46378082 1.32698675## [7] -1.12412636 1.20387148 2.82675459 0.25252621 0.58829513 -0.25112717## [13] -0.13920420 0.08464175 0.07344945 -0.01608893 -0.23993487 0.90727560## [19] 0.37564148 1.14790999 1.20946763 -0.54772305 -0.30708866 -1.36476075## [25] -0.44699237 0.58829513 -0.64285758 -0.53093460 -1.87401028 -1.31439542## [31] -1.81804880 0.42041067## ## $vs## [1] -0.8680278 -0.8680278 1.1160357 1.1160357 -0.8680278 1.1160357 -0.8680278## [8] 1.1160357 1.1160357 1.1160357 1.1160357 -0.8680278 -0.8680278 -0.8680278## [15] -0.8680278 -0.8680278 -0.8680278 1.1160357 1.1160357 1.1160357 1.1160357## [22] -0.8680278 -0.8680278 -0.8680278 -0.8680278 1.1160357 -0.8680278 1.1160357## [29] -0.8680278 -0.8680278 -0.8680278 1.1160357## ## $am## [1] 1.1899014 1.1899014 1.1899014 -0.8141431 -0.8141431 -0.8141431 -0.8141431## [8] -0.8141431 -0.8141431 -0.8141431 -0.8141431 -0.8141431 -0.8141431 -0.8141431## [15] -0.8141431 -0.8141431 -0.8141431 1.1899014 1.1899014 1.1899014 -0.8141431## [22] -0.8141431 -0.8141431 -0.8141431 -0.8141431 1.1899014 1.1899014 1.1899014## [29] 1.1899014 1.1899014 1.1899014 1.1899014## ## $gear## [1] 0.4235542 0.4235542 0.4235542 -0.9318192 -0.9318192 -0.9318192 -0.9318192## [8] 0.4235542 0.4235542 0.4235542 0.4235542 -0.9318192 -0.9318192 -0.9318192## [15] -0.9318192 -0.9318192 -0.9318192 0.4235542 0.4235542 0.4235542 -0.9318192## [22] -0.9318192 -0.9318192 -0.9318192 -0.9318192 0.4235542 1.7789276 1.7789276## [29] 1.7789276 1.7789276 1.7789276 0.4235542## ## $carb## [1] 0.7352031 0.7352031 -1.1221521 -1.1221521 -0.5030337 -1.1221521 0.7352031## [8] -0.5030337 -0.5030337 0.7352031 0.7352031 0.1160847 0.1160847 0.1160847## [15] 0.7352031 0.7352031 0.7352031 -1.1221521 -0.5030337 -1.1221521 -1.1221521## [22] -0.5030337 -0.5030337 0.7352031 -0.5030337 -1.1221521 -0.5030337 -0.5030337## [29] 0.7352031 1.9734398 3.2116766 -0.5030337modify
modify(mtcars, ~as.numeric(scale(.x)))## mpg cyl disp hp drat## Mazda RX4 0.15088482 -0.1049878 -0.57061982 -0.53509284 0.56751369## Mazda RX4 Wag 0.15088482 -0.1049878 -0.57061982 -0.53509284 0.56751369## Datsun 710 0.44954345 -1.2248578 -0.99018209 -0.78304046 0.47399959## Hornet 4 Drive 0.21725341 -0.1049878 0.22009369 -0.53509284 -0.96611753## Hornet Sportabout -0.23073453 1.0148821 1.04308123 0.41294217 -0.83519779## Valiant -0.33028740 -0.1049878 -0.04616698 -0.60801861 -1.56460776## Duster 360 -0.96078893 1.0148821 1.04308123 1.43390296 -0.72298087## Merc 240D 0.71501778 -1.2248578 -0.67793094 -1.23518023 0.17475447## Merc 230 0.44954345 -1.2248578 -0.72553512 -0.75387015 0.60491932## Merc 280 -0.14777380 -0.1049878 -0.50929918 -0.34548584 0.60491932## Merc 280C -0.38006384 -0.1049878 -0.50929918 -0.34548584 0.60491932## Merc 450SE -0.61235388 1.0148821 0.36371309 0.48586794 -0.98482035## Merc 450SL -0.46302456 1.0148821 0.36371309 0.48586794 -0.98482035## Merc 450SLC -0.81145962 1.0148821 0.36371309 0.48586794 -0.98482035## Cadillac Fleetwood -1.60788262 1.0148821 1.94675381 0.85049680 -1.24665983## Lincoln Continental -1.60788262 1.0148821 1.84993175 0.99634834 -1.11574009## Chrysler Imperial -0.89442035 1.0148821 1.68856165 1.21512565 -0.68557523## Fiat 128 2.04238943 -1.2248578 -1.22658929 -1.17683962 0.90416444## Honda Civic 1.71054652 -1.2248578 -1.25079481 -1.38103178 2.49390411## Toyota Corolla 2.29127162 -1.2248578 -1.28790993 -1.19142477 1.16600392## Toyota Corona 0.23384555 -1.2248578 -0.89255318 -0.72469984 0.19345729## Dodge Challenger -0.76168319 1.0148821 0.70420401 0.04831332 -1.56460776## AMC Javelin -0.81145962 1.0148821 0.59124494 0.04831332 -0.83519779## Camaro Z28 -1.12671039 1.0148821 0.96239618 1.43390296 0.24956575## Pontiac Firebird -0.14777380 1.0148821 1.36582144 0.41294217 -0.96611753## Fiat X1-9 1.19619000 -1.2248578 -1.22416874 -1.17683962 0.90416444## Porsche 914-2 0.98049211 -1.2248578 -0.89093948 -0.81221077 1.55876313## Lotus Europa 1.71054652 -1.2248578 -1.09426581 -0.49133738 0.32437703## Ford Pantera L -0.71190675 1.0148821 0.97046468 1.71102089 1.16600392## Ferrari Dino -0.06481307 -0.1049878 -0.69164740 0.41294217 0.04383473## Maserati Bora -0.84464392 1.0148821 0.56703942 2.74656682 -0.10578782## Volvo 142E 0.21725341 -1.2248578 -0.88529152 -0.54967799 0.96027290## wt qsec vs am gear## Mazda RX4 -0.610399567 -0.77716515 -0.8680278 1.1899014 0.4235542## Mazda RX4 Wag -0.349785269 -0.46378082 -0.8680278 1.1899014 0.4235542## Datsun 710 -0.917004624 0.42600682 1.1160357 1.1899014 0.4235542## Hornet 4 Drive -0.002299538 0.89048716 1.1160357 -0.8141431 -0.9318192## Hornet Sportabout 0.227654255 -0.46378082 -0.8680278 -0.8141431 -0.9318192## Valiant 0.248094592 1.32698675 1.1160357 -0.8141431 -0.9318192## Duster 360 0.360516446 -1.12412636 -0.8680278 -0.8141431 -0.9318192## Merc 240D -0.027849959 1.20387148 1.1160357 -0.8141431 0.4235542## Merc 230 -0.068730634 2.82675459 1.1160357 -0.8141431 0.4235542## Merc 280 0.227654255 0.25252621 1.1160357 -0.8141431 0.4235542## Merc 280C 0.227654255 0.58829513 1.1160357 -0.8141431 0.4235542## Merc 450SE 0.871524874 -0.25112717 -0.8680278 -0.8141431 -0.9318192## Merc 450SL 0.524039143 -0.13920420 -0.8680278 -0.8141431 -0.9318192## Merc 450SLC 0.575139986 0.08464175 -0.8680278 -0.8141431 -0.9318192## Cadillac Fleetwood 2.077504765 0.07344945 -0.8680278 -0.8141431 -0.9318192## Lincoln Continental 2.255335698 -0.01608893 -0.8680278 -0.8141431 -0.9318192## Chrysler Imperial 2.174596366 -0.23993487 -0.8680278 -0.8141431 -0.9318192## Fiat 128 -1.039646647 0.90727560 1.1160357 1.1899014 0.4235542## Honda Civic -1.637526508 0.37564148 1.1160357 1.1899014 0.4235542## Toyota Corolla -1.412682800 1.14790999 1.1160357 1.1899014 0.4235542## Toyota Corona -0.768812180 1.20946763 1.1160357 -0.8141431 -0.9318192## Dodge Challenger 0.309415603 -0.54772305 -0.8680278 -0.8141431 -0.9318192## AMC Javelin 0.222544170 -0.30708866 -0.8680278 -0.8141431 -0.9318192## Camaro Z28 0.636460997 -1.36476075 -0.8680278 -0.8141431 -0.9318192## Pontiac Firebird 0.641571082 -0.44699237 -0.8680278 -0.8141431 -0.9318192## Fiat X1-9 -1.310481114 0.58829513 1.1160357 1.1899014 0.4235542## Porsche 914-2 -1.100967659 -0.64285758 -0.8680278 1.1899014 1.7789276## Lotus Europa -1.741772228 -0.53093460 1.1160357 1.1899014 1.7789276## Ford Pantera L -0.048290296 -1.87401028 -0.8680278 1.1899014 1.7789276## Ferrari Dino -0.457097039 -1.31439542 -0.8680278 1.1899014 1.7789276## Maserati Bora 0.360516446 -1.81804880 -0.8680278 1.1899014 1.7789276## Volvo 142E -0.446876870 0.42041067 1.1160357 1.1899014 0.4235542## carb## Mazda RX4 0.7352031## Mazda RX4 Wag 0.7352031## Datsun 710 -1.1221521## Hornet 4 Drive -1.1221521## Hornet Sportabout -0.5030337## Valiant -1.1221521## Duster 360 0.7352031## Merc 240D -0.5030337## Merc 230 -0.5030337## Merc 280 0.7352031## Merc 280C 0.7352031## Merc 450SE 0.1160847## Merc 450SL 0.1160847## Merc 450SLC 0.1160847## Cadillac Fleetwood 0.7352031## Lincoln Continental 0.7352031## Chrysler Imperial 0.7352031## Fiat 128 -1.1221521## Honda Civic -0.5030337## Toyota Corolla -1.1221521## Toyota Corona -1.1221521## Dodge Challenger -0.5030337## AMC Javelin -0.5030337## Camaro Z28 0.7352031## Pontiac Firebird -0.5030337## Fiat X1-9 -1.1221521## Porsche 914-2 -0.5030337## Lotus Europa -0.5030337## Ford Pantera L 0.7352031## Ferrari Dino 1.9734398## Maserati Bora 3.2116766## Volvo 142E -0.5030337Try to fit a model
(please follow along)
Notice the error message is super helpful! (this is new)
by_cyl %>% mutate(mod = map(data, ~lm(hwy ~ displ + drv, data = .x)))## Error in `mutate()`:## ! Problem while computing `mod = map(data, ~lm(hwy ~ displ +## drv, data = .x))`.## ℹ The error occurred in group 2: cyl = 5.## Caused by error in `contrasts<-`:## ! contrasts can be applied only to factors with 2 or more levelsSafe return
- First, define safe function - note that this will work for any function
safe_lm <- safely(lm)- Next, loop the safe function, instead of the standard function
safe_models <- by_cyl %>% mutate(safe_mod = map(data, ~safe_lm(hwy ~ displ + drv, data = .x)))safe_models## # A tibble: 4 × 3## # Groups: cyl [4]## cyl data safe_mod ## <int> <list> <list> ## 1 4 <tibble [81 × 10]> <named list [2]>## 2 6 <tibble [79 × 10]> <named list [2]>## 3 8 <tibble [70 × 10]> <named list [2]>## 4 5 <tibble [4 × 10]> <named list [2]>What's returned?
safe_models$safe_mod[[1]]## $result## ## Call:## .f(formula = ..1, data = ..2)## ## Coefficients:## (Intercept) displ drvf ## 37.370 -5.289 3.882 ## ## ## $error## NULLsafe_models$safe_mod[[4]]## $result## NULL## ## $error## <simpleError in `contrasts<-`(`*tmp*`, value = contr.funs[1 + isOF[nn]]): contrasts can be applied only to factors with 2 or more levels>Inspecting
I often use safely() to help me de-bug. Why is it failing there (but note the new error messages help with this too).
First - create a new variable to filter for results with errors
safe_models %>% mutate(error = map_lgl(safe_mod, ~!is.null(.x$error)))## # A tibble: 4 × 4## # Groups: cyl [4]## cyl data safe_mod error## <int> <list> <list> <lgl>## 1 4 <tibble [81 × 10]> <named list [2]> FALSE## 2 6 <tibble [79 × 10]> <named list [2]> FALSE## 3 8 <tibble [70 × 10]> <named list [2]> FALSE## 4 5 <tibble [4 × 10]> <named list [2]> TRUEInspecting the data
safe_models %>% mutate(error = map_lgl(safe_mod, ~!is.null(.x$error))) %>% filter(isTRUE(error)) %>% select(cyl, data) %>% unnest(data)## # A tibble: 4 × 11## # Groups: cyl [1]## cyl manufacturer model displ year trans drv cty hwy fl ## <int> <chr> <chr> <dbl> <int> <chr> <chr> <int> <int> <chr>## 1 5 volkswagen jetta 2.5 2008 auto(s6) f 21 29 r ## 2 5 volkswagen jetta 2.5 2008 manual(m5) f 21 29 r ## 3 5 volkswagen new beetle 2.5 2008 manual(m5) f 20 28 r ## 4 5 volkswagen new beetle 2.5 2008 auto(s6) f 20 29 r ## # … with 1 more variable: class <chr>The displ and drv variables are constant, so no relation can be estimated.
Pull results that worked
safe_models %>% mutate(results = map(safe_mod, "result"))## # A tibble: 4 × 4## # Groups: cyl [4]## cyl data safe_mod results## <int> <list> <list> <list> ## 1 4 <tibble [81 × 10]> <named list [2]> <lm> ## 2 6 <tibble [79 × 10]> <named list [2]> <lm> ## 3 8 <tibble [70 × 10]> <named list [2]> <lm> ## 4 5 <tibble [4 × 10]> <named list [2]> <NULL>Pull results that worked
safe_models %>% mutate(results = map(safe_mod, "result"))## # A tibble: 4 × 4## # Groups: cyl [4]## cyl data safe_mod results## <int> <list> <list> <list> ## 1 4 <tibble [81 × 10]> <named list [2]> <lm> ## 2 6 <tibble [79 × 10]> <named list [2]> <lm> ## 3 8 <tibble [70 × 10]> <named list [2]> <lm> ## 4 5 <tibble [4 × 10]> <named list [2]> <NULL>Now we can broom::tidy() or whatevs
Notice that there is no cyl == 5.
safe_models %>% mutate(results = map(safe_mod, "result"), tidied = map(results, broom::tidy)) %>% select(cyl, tidied) %>% unnest(tidied)## # A tibble: 11 × 6## # Groups: cyl [3]## cyl term estimate std.error statistic p.value## <int> <chr> <dbl> <dbl> <dbl> <dbl>## 1 4 (Intercept) 37.37023 3.537572 10.56381 1.052943e-16## 2 4 displ -5.288562 1.436068 -3.682668 4.235795e- 4## 3 4 drvf 3.882134 0.9971876 3.893083 2.073699e- 4## 4 6 (Intercept) 27.96536 2.347630 11.91217 5.718039e-19## 5 6 displ -2.333261 0.6373304 -3.660991 4.651570e- 4## 6 6 drvf 4.570840 0.6012367 7.602397 6.789988e-11## 7 6 drvr 6.384355 1.229277 5.193585 1.713129e- 6## 8 8 (Intercept) 14.82265 2.887289 5.133759 2.708515e- 6## 9 8 displ 0.3060487 0.5719058 0.5351383 5.943528e- 1## 10 8 drvf 8.555294 2.679129 3.193311 2.156229e- 3## 11 8 drvr 3.709336 0.7319048 5.068058 3.473594e- 6Example
library(rvest)links <- list( "https://en.wikipedia.org/wiki/FC_Barcelona", "https://nosuchpage", "https://en.wikipedia.org/wiki/Rome")pages <- map(links, ~{ Sys.sleep(0.1) read_html(.x)})## Error in open.connection(x, "rb"): Timeout was reached: [nosuchpage] Failed to connect to nosuchpage port 443: Operation timed outThe problem
I can't connect to https://nosuchpage because it doesn't exist
BUT
That also means I can't get any of my links because one page errored (imagine it was 1 in 1,000 instead of 1 in 3)
The problem
I can't connect to https://nosuchpage because it doesn't exist
BUT
That also means I can't get any of my links because one page errored (imagine it was 1 in 1,000 instead of 1 in 3)
safely() to the rescue
Safe version
safe_read_html <- safely(read_html)pages <- map(links, ~{ Sys.sleep(0.1) safe_read_html(.x)})str(pages)## List of 3## $ :List of 2## ..$ result:List of 2## .. ..$ node:<externalptr> ## .. ..$ doc :<externalptr> ## .. ..- attr(*, "class")= chr [1:2] "xml_document" "xml_node"## ..$ error : NULL## $ :List of 2## ..$ result: NULL## ..$ error :List of 2## .. ..$ message: chr "Failed to connect to nosuchpage port 443: Connection refused"## .. ..$ call : language open.connection(x, "rb")## .. ..- attr(*, "class")= chr [1:3] "simpleError" "error" "condition"## $ :List of 2## ..$ result:List of 2## .. ..$ node:<externalptr> ## .. ..$ doc :<externalptr> ## .. ..- attr(*, "class")= chr [1:2] "xml_document" "xml_node"## ..$ error : NULLWhy might you use this?
What if you had a list of data frames like this
l_df <- list( tibble(id = 1:3, score = rnorm(3)), tibble(id = 1:5, treatment = rbinom(5, 1, .5)), tibble(id = c(1, 3, 5, 7), other_thing = rnorm(4)))We can join these all together with a single loop - we want the output to be of length 1!
Note - you have to be careful on directionality
reduce(l_df, left_join)## # A tibble: 3 × 4## id score treatment other_thing## <dbl> <dbl> <int> <dbl>## 1 1 0.2548487 0 0.8905293## 2 2 1.272697 0 NA ## 3 3 -0.7417612 1 0.5923346reduce(l_df, right_join)## # A tibble: 4 × 4## id score treatment other_thing## <dbl> <dbl> <int> <dbl>## 1 1 0.2548487 0 0.8905293## 2 3 -0.7417612 1 0.5923346## 3 5 NA 0 0.4740725## 4 7 NA NA -0.1488833Another example
You probably just want to bind_rows()
l_df2 <- list( tibble(id = 1:3, scid = 1, score = rnorm(3)), tibble(id = 1:5, scid = 2, score = rnorm(5)), tibble(id = c(1, 3, 5, 7), scid = 3, score = rnorm(4)))reduce(l_df2, bind_rows)## # A tibble: 12 × 3## id scid score## <dbl> <dbl> <dbl>## 1 1 1 1.636123 ## 2 2 1 0.9520929## 3 3 1 -1.635054 ## 4 1 2 -1.595450 ## 5 2 2 0.3252145## 6 3 2 0.9735523## 7 4 2 -0.9043736## 8 5 2 -0.4567605## 9 1 3 -1.208666 ## 10 3 3 -2.079515 ## 11 5 3 -0.7194680## 12 7 3 1.207981Non-loop version
Luckily, the prior slide has become obsolete, because bind_rows() will do the list reduction for us.
bind_rows(l_df2)## # A tibble: 12 × 3## id scid score## <dbl> <dbl> <dbl>## 1 1 1 1.636123 ## 2 2 1 0.9520929## 3 3 1 -1.635054 ## 4 1 2 -1.595450 ## 5 2 2 0.3252145## 6 3 2 0.9735523## 7 4 2 -0.9043736## 8 5 2 -0.4567605## 9 1 3 -1.208666 ## 10 3 3 -2.079515 ## 11 5 3 -0.7194680## 12 7 3 1.207981Another example
This is a poor example, but there are use cases like this
library(palmerpenguins)map(penguins, as.character) %>% reduce(paste)## [1] "Adelie Torgersen 39.1 18.7 181 3750 male 2007" ## [2] "Adelie Torgersen 39.5 17.4 186 3800 female 2007"## [3] "Adelie Torgersen 40.3 18 195 3250 female 2007" ## [4] "Adelie Torgersen NA NA NA NA NA 2007" ## [5] "Adelie Torgersen 36.7 19.3 193 3450 female 2007"## [6] "Adelie Torgersen 39.3 20.6 190 3650 male 2007" ## [7] "Adelie Torgersen 38.9 17.8 181 3625 female 2007"## [8] "Adelie Torgersen 39.2 19.6 195 4675 male 2007" ## [9] "Adelie Torgersen 34.1 18.1 193 3475 NA 2007" ## [10] "Adelie Torgersen 42 20.2 190 4250 NA 2007" ## [11] "Adelie Torgersen 37.8 17.1 186 3300 NA 2007" ## [12] "Adelie Torgersen 37.8 17.3 180 3700 NA 2007" ## [13] "Adelie Torgersen 41.1 17.6 182 3200 female 2007"## [14] "Adelie Torgersen 38.6 21.2 191 3800 male 2007" ## [15] "Adelie Torgersen 34.6 21.1 198 4400 male 2007" ## [16] "Adelie Torgersen 36.6 17.8 185 3700 female 2007"## [17] "Adelie Torgersen 38.7 19 195 3450 female 2007" ## [18] "Adelie Torgersen 42.5 20.7 197 4500 male 2007" ## [19] "Adelie Torgersen 34.4 18.4 184 3325 female 2007"## [20] "Adelie Torgersen 46 21.5 194 4200 male 2007" ## [21] "Adelie Biscoe 37.8 18.3 174 3400 female 2007" ## [22] "Adelie Biscoe 37.7 18.7 180 3600 male 2007" ## [23] "Adelie Biscoe 35.9 19.2 189 3800 female 2007" ## [24] "Adelie Biscoe 38.2 18.1 185 3950 male 2007" ## [25] "Adelie Biscoe 38.8 17.2 180 3800 male 2007" ## [26] "Adelie Biscoe 35.3 18.9 187 3800 female 2007" ## [27] "Adelie Biscoe 40.6 18.6 183 3550 male 2007" ## [28] "Adelie Biscoe 40.5 17.9 187 3200 female 2007" ## [29] "Adelie Biscoe 37.9 18.6 172 3150 female 2007" ## [30] "Adelie Biscoe 40.5 18.9 180 3950 male 2007" ## [31] "Adelie Dream 39.5 16.7 178 3250 female 2007" ## [32] "Adelie Dream 37.2 18.1 178 3900 male 2007" ## [33] "Adelie Dream 39.5 17.8 188 3300 female 2007" ## [34] "Adelie Dream 40.9 18.9 184 3900 male 2007" ## [35] "Adelie Dream 36.4 17 195 3325 female 2007" ## [36] "Adelie Dream 39.2 21.1 196 4150 male 2007" ## [37] "Adelie Dream 38.8 20 190 3950 male 2007" ## [38] "Adelie Dream 42.2 18.5 180 3550 female 2007" ## [39] "Adelie Dream 37.6 19.3 181 3300 female 2007" ## [40] "Adelie Dream 39.8 19.1 184 4650 male 2007" ## [41] "Adelie Dream 36.5 18 182 3150 female 2007" ## [42] "Adelie Dream 40.8 18.4 195 3900 male 2007" ## [43] "Adelie Dream 36 18.5 186 3100 female 2007" ## [44] "Adelie Dream 44.1 19.7 196 4400 male 2007" ## [45] "Adelie Dream 37 16.9 185 3000 female 2007" ## [46] "Adelie Dream 39.6 18.8 190 4600 male 2007" ## [47] "Adelie Dream 41.1 19 182 3425 male 2007" ## [48] "Adelie Dream 37.5 18.9 179 2975 NA 2007" ## [49] "Adelie Dream 36 17.9 190 3450 female 2007" ## [50] "Adelie Dream 42.3 21.2 191 4150 male 2007" ## [51] "Adelie Biscoe 39.6 17.7 186 3500 female 2008" ## [52] "Adelie Biscoe 40.1 18.9 188 4300 male 2008" ## [53] "Adelie Biscoe 35 17.9 190 3450 female 2008" ## [54] "Adelie Biscoe 42 19.5 200 4050 male 2008" ## [55] "Adelie Biscoe 34.5 18.1 187 2900 female 2008" ## [56] "Adelie Biscoe 41.4 18.6 191 3700 male 2008" ## [57] "Adelie Biscoe 39 17.5 186 3550 female 2008" ## [58] "Adelie Biscoe 40.6 18.8 193 3800 male 2008" ## [59] "Adelie Biscoe 36.5 16.6 181 2850 female 2008" ## [60] "Adelie Biscoe 37.6 19.1 194 3750 male 2008" ## [61] "Adelie Biscoe 35.7 16.9 185 3150 female 2008" ## [62] "Adelie Biscoe 41.3 21.1 195 4400 male 2008" ## [63] "Adelie Biscoe 37.6 17 185 3600 female 2008" ## [64] "Adelie Biscoe 41.1 18.2 192 4050 male 2008" ## [65] "Adelie Biscoe 36.4 17.1 184 2850 female 2008" ## [66] "Adelie Biscoe 41.6 18 192 3950 male 2008" ## [67] "Adelie Biscoe 35.5 16.2 195 3350 female 2008" ## [68] "Adelie Biscoe 41.1 19.1 188 4100 male 2008" ## [69] "Adelie Torgersen 35.9 16.6 190 3050 female 2008"## [70] "Adelie Torgersen 41.8 19.4 198 4450 male 2008" ## [71] "Adelie Torgersen 33.5 19 190 3600 female 2008" ## [72] "Adelie Torgersen 39.7 18.4 190 3900 male 2008" ## [73] "Adelie Torgersen 39.6 17.2 196 3550 female 2008"## [74] "Adelie Torgersen 45.8 18.9 197 4150 male 2008" ## [75] "Adelie Torgersen 35.5 17.5 190 3700 female 2008"## [76] "Adelie Torgersen 42.8 18.5 195 4250 male 2008" ## [77] "Adelie Torgersen 40.9 16.8 191 3700 female 2008"## [78] "Adelie Torgersen 37.2 19.4 184 3900 male 2008" ## [79] "Adelie Torgersen 36.2 16.1 187 3550 female 2008"## [80] "Adelie Torgersen 42.1 19.1 195 4000 male 2008" ## [81] "Adelie Torgersen 34.6 17.2 189 3200 female 2008"## [82] "Adelie Torgersen 42.9 17.6 196 4700 male 2008" ## [83] "Adelie Torgersen 36.7 18.8 187 3800 female 2008"## [84] "Adelie Torgersen 35.1 19.4 193 4200 male 2008" ## [85] "Adelie Dream 37.3 17.8 191 3350 female 2008" ## [86] "Adelie Dream 41.3 20.3 194 3550 male 2008" ## [87] "Adelie Dream 36.3 19.5 190 3800 male 2008" ## [88] "Adelie Dream 36.9 18.6 189 3500 female 2008" ## [89] "Adelie Dream 38.3 19.2 189 3950 male 2008" ## [90] "Adelie Dream 38.9 18.8 190 3600 female 2008" ## [91] "Adelie Dream 35.7 18 202 3550 female 2008" ## [92] "Adelie Dream 41.1 18.1 205 4300 male 2008" ## [93] "Adelie Dream 34 17.1 185 3400 female 2008" ## [94] "Adelie Dream 39.6 18.1 186 4450 male 2008" ## [95] "Adelie Dream 36.2 17.3 187 3300 female 2008" ## [96] "Adelie Dream 40.8 18.9 208 4300 male 2008" ## [97] "Adelie Dream 38.1 18.6 190 3700 female 2008" ## [98] "Adelie Dream 40.3 18.5 196 4350 male 2008" ## [99] "Adelie Dream 33.1 16.1 178 2900 female 2008" ## [100] "Adelie Dream 43.2 18.5 192 4100 male 2008" ## [101] "Adelie Biscoe 35 17.9 192 3725 female 2009" ## [102] "Adelie Biscoe 41 20 203 4725 male 2009" ## [103] "Adelie Biscoe 37.7 16 183 3075 female 2009" ## [104] "Adelie Biscoe 37.8 20 190 4250 male 2009" ## [105] "Adelie Biscoe 37.9 18.6 193 2925 female 2009" ## [106] "Adelie Biscoe 39.7 18.9 184 3550 male 2009" ## [107] "Adelie Biscoe 38.6 17.2 199 3750 female 2009" ## [108] "Adelie Biscoe 38.2 20 190 3900 male 2009" ## [109] "Adelie Biscoe 38.1 17 181 3175 female 2009" ## [110] "Adelie Biscoe 43.2 19 197 4775 male 2009" ## [111] "Adelie Biscoe 38.1 16.5 198 3825 female 2009" ## [112] "Adelie Biscoe 45.6 20.3 191 4600 male 2009" ## [113] "Adelie Biscoe 39.7 17.7 193 3200 female 2009" ## [114] "Adelie Biscoe 42.2 19.5 197 4275 male 2009" ## [115] "Adelie Biscoe 39.6 20.7 191 3900 female 2009" ## [116] "Adelie Biscoe 42.7 18.3 196 4075 male 2009" ## [117] "Adelie Torgersen 38.6 17 188 2900 female 2009" ## [118] "Adelie Torgersen 37.3 20.5 199 3775 male 2009" ## [119] "Adelie Torgersen 35.7 17 189 3350 female 2009" ## [120] "Adelie Torgersen 41.1 18.6 189 3325 male 2009" ## [121] "Adelie Torgersen 36.2 17.2 187 3150 female 2009"## [122] "Adelie Torgersen 37.7 19.8 198 3500 male 2009" ## [123] "Adelie Torgersen 40.2 17 176 3450 female 2009" ## [124] "Adelie Torgersen 41.4 18.5 202 3875 male 2009" ## [125] "Adelie Torgersen 35.2 15.9 186 3050 female 2009"## [126] "Adelie Torgersen 40.6 19 199 4000 male 2009" ## [127] "Adelie Torgersen 38.8 17.6 191 3275 female 2009"## [128] "Adelie Torgersen 41.5 18.3 195 4300 male 2009" ## [129] "Adelie Torgersen 39 17.1 191 3050 female 2009" ## [130] "Adelie Torgersen 44.1 18 210 4000 male 2009" ## [131] "Adelie Torgersen 38.5 17.9 190 3325 female 2009"## [132] "Adelie Torgersen 43.1 19.2 197 3500 male 2009" ## [133] "Adelie Dream 36.8 18.5 193 3500 female 2009" ## [134] "Adelie Dream 37.5 18.5 199 4475 male 2009" ## [135] "Adelie Dream 38.1 17.6 187 3425 female 2009" ## [136] "Adelie Dream 41.1 17.5 190 3900 male 2009" ## [137] "Adelie Dream 35.6 17.5 191 3175 female 2009" ## [138] "Adelie Dream 40.2 20.1 200 3975 male 2009" ## [139] "Adelie Dream 37 16.5 185 3400 female 2009" ## [140] "Adelie Dream 39.7 17.9 193 4250 male 2009" ## [141] "Adelie Dream 40.2 17.1 193 3400 female 2009" ## [142] "Adelie Dream 40.6 17.2 187 3475 male 2009" ## [143] "Adelie Dream 32.1 15.5 188 3050 female 2009" ## [144] "Adelie Dream 40.7 17 190 3725 male 2009" ## [145] "Adelie Dream 37.3 16.8 192 3000 female 2009" ## [146] "Adelie Dream 39 18.7 185 3650 male 2009" ## [147] "Adelie Dream 39.2 18.6 190 4250 male 2009" ## [148] "Adelie Dream 36.6 18.4 184 3475 female 2009" ## [149] "Adelie Dream 36 17.8 195 3450 female 2009" ## [150] "Adelie Dream 37.8 18.1 193 3750 male 2009" ## [151] "Adelie Dream 36 17.1 187 3700 female 2009" ## [152] "Adelie Dream 41.5 18.5 201 4000 male 2009" ## [153] "Gentoo Biscoe 46.1 13.2 211 4500 female 2007" ## [154] "Gentoo Biscoe 50 16.3 230 5700 male 2007" ## [155] "Gentoo Biscoe 48.7 14.1 210 4450 female 2007" ## [156] "Gentoo Biscoe 50 15.2 218 5700 male 2007" ## [157] "Gentoo Biscoe 47.6 14.5 215 5400 male 2007" ## [158] "Gentoo Biscoe 46.5 13.5 210 4550 female 2007" ## [159] "Gentoo Biscoe 45.4 14.6 211 4800 female 2007" ## [160] "Gentoo Biscoe 46.7 15.3 219 5200 male 2007" ## [161] "Gentoo Biscoe 43.3 13.4 209 4400 female 2007" ## [162] "Gentoo Biscoe 46.8 15.4 215 5150 male 2007" ## [163] "Gentoo Biscoe 40.9 13.7 214 4650 female 2007" ## [164] "Gentoo Biscoe 49 16.1 216 5550 male 2007" ## [165] "Gentoo Biscoe 45.5 13.7 214 4650 female 2007" ## [166] "Gentoo Biscoe 48.4 14.6 213 5850 male 2007" ## [167] "Gentoo Biscoe 45.8 14.6 210 4200 female 2007" ## [168] "Gentoo Biscoe 49.3 15.7 217 5850 male 2007" ## [169] "Gentoo Biscoe 42 13.5 210 4150 female 2007" ## [170] "Gentoo Biscoe 49.2 15.2 221 6300 male 2007" ## [171] "Gentoo Biscoe 46.2 14.5 209 4800 female 2007" ## [172] "Gentoo Biscoe 48.7 15.1 222 5350 male 2007" ## [173] "Gentoo Biscoe 50.2 14.3 218 5700 male 2007" ## [174] "Gentoo Biscoe 45.1 14.5 215 5000 female 2007" ## [175] "Gentoo Biscoe 46.5 14.5 213 4400 female 2007" ## [176] "Gentoo Biscoe 46.3 15.8 215 5050 male 2007" ## [177] "Gentoo Biscoe 42.9 13.1 215 5000 female 2007" ## [178] "Gentoo Biscoe 46.1 15.1 215 5100 male 2007" ## [179] "Gentoo Biscoe 44.5 14.3 216 4100 NA 2007" ## [180] "Gentoo Biscoe 47.8 15 215 5650 male 2007" ## [181] "Gentoo Biscoe 48.2 14.3 210 4600 female 2007" ## [182] "Gentoo Biscoe 50 15.3 220 5550 male 2007" ## [183] "Gentoo Biscoe 47.3 15.3 222 5250 male 2007" ## [184] "Gentoo Biscoe 42.8 14.2 209 4700 female 2007" ## [185] "Gentoo Biscoe 45.1 14.5 207 5050 female 2007" ## [186] "Gentoo Biscoe 59.6 17 230 6050 male 2007" ## [187] "Gentoo Biscoe 49.1 14.8 220 5150 female 2008" ## [188] "Gentoo Biscoe 48.4 16.3 220 5400 male 2008" ## [189] "Gentoo Biscoe 42.6 13.7 213 4950 female 2008" ## [190] "Gentoo Biscoe 44.4 17.3 219 5250 male 2008" ## [191] "Gentoo Biscoe 44 13.6 208 4350 female 2008" ## [192] "Gentoo Biscoe 48.7 15.7 208 5350 male 2008" ## [193] "Gentoo Biscoe 42.7 13.7 208 3950 female 2008" ## [194] "Gentoo Biscoe 49.6 16 225 5700 male 2008" ## [195] "Gentoo Biscoe 45.3 13.7 210 4300 female 2008" ## [196] "Gentoo Biscoe 49.6 15 216 4750 male 2008" ## [197] "Gentoo Biscoe 50.5 15.9 222 5550 male 2008" ## [198] "Gentoo Biscoe 43.6 13.9 217 4900 female 2008" ## [199] "Gentoo Biscoe 45.5 13.9 210 4200 female 2008" ## [200] "Gentoo Biscoe 50.5 15.9 225 5400 male 2008" ## [201] "Gentoo Biscoe 44.9 13.3 213 5100 female 2008" ## [202] "Gentoo Biscoe 45.2 15.8 215 5300 male 2008" ## [203] "Gentoo Biscoe 46.6 14.2 210 4850 female 2008" ## [204] "Gentoo Biscoe 48.5 14.1 220 5300 male 2008" ## [205] "Gentoo Biscoe 45.1 14.4 210 4400 female 2008" ## [206] "Gentoo Biscoe 50.1 15 225 5000 male 2008" ## [207] "Gentoo Biscoe 46.5 14.4 217 4900 female 2008" ## [208] "Gentoo Biscoe 45 15.4 220 5050 male 2008" ## [209] "Gentoo Biscoe 43.8 13.9 208 4300 female 2008" ## [210] "Gentoo Biscoe 45.5 15 220 5000 male 2008" ## [211] "Gentoo Biscoe 43.2 14.5 208 4450 female 2008" ## [212] "Gentoo Biscoe 50.4 15.3 224 5550 male 2008" ## [213] "Gentoo Biscoe 45.3 13.8 208 4200 female 2008" ## [214] "Gentoo Biscoe 46.2 14.9 221 5300 male 2008" ## [215] "Gentoo Biscoe 45.7 13.9 214 4400 female 2008" ## [216] "Gentoo Biscoe 54.3 15.7 231 5650 male 2008" ## [217] "Gentoo Biscoe 45.8 14.2 219 4700 female 2008" ## [218] "Gentoo Biscoe 49.8 16.8 230 5700 male 2008" ## [219] "Gentoo Biscoe 46.2 14.4 214 4650 NA 2008" ## [220] "Gentoo Biscoe 49.5 16.2 229 5800 male 2008" ## [221] "Gentoo Biscoe 43.5 14.2 220 4700 female 2008" ## [222] "Gentoo Biscoe 50.7 15 223 5550 male 2008" ## [223] "Gentoo Biscoe 47.7 15 216 4750 female 2008" ## [224] "Gentoo Biscoe 46.4 15.6 221 5000 male 2008" ## [225] "Gentoo Biscoe 48.2 15.6 221 5100 male 2008" ## [226] "Gentoo Biscoe 46.5 14.8 217 5200 female 2008" ## [227] "Gentoo Biscoe 46.4 15 216 4700 female 2008" ## [228] "Gentoo Biscoe 48.6 16 230 5800 male 2008" ## [229] "Gentoo Biscoe 47.5 14.2 209 4600 female 2008" ## [230] "Gentoo Biscoe 51.1 16.3 220 6000 male 2008" ## [231] "Gentoo Biscoe 45.2 13.8 215 4750 female 2008" ## [232] "Gentoo Biscoe 45.2 16.4 223 5950 male 2008" ## [233] "Gentoo Biscoe 49.1 14.5 212 4625 female 2009" ## [234] "Gentoo Biscoe 52.5 15.6 221 5450 male 2009" ## [235] "Gentoo Biscoe 47.4 14.6 212 4725 female 2009" ## [236] "Gentoo Biscoe 50 15.9 224 5350 male 2009" ## [237] "Gentoo Biscoe 44.9 13.8 212 4750 female 2009" ## [238] "Gentoo Biscoe 50.8 17.3 228 5600 male 2009" ## [239] "Gentoo Biscoe 43.4 14.4 218 4600 female 2009" ## [240] "Gentoo Biscoe 51.3 14.2 218 5300 male 2009" ## [241] "Gentoo Biscoe 47.5 14 212 4875 female 2009" ## [242] "Gentoo Biscoe 52.1 17 230 5550 male 2009" ## [243] "Gentoo Biscoe 47.5 15 218 4950 female 2009" ## [244] "Gentoo Biscoe 52.2 17.1 228 5400 male 2009" ## [245] "Gentoo Biscoe 45.5 14.5 212 4750 female 2009" ## [246] "Gentoo Biscoe 49.5 16.1 224 5650 male 2009" ## [247] "Gentoo Biscoe 44.5 14.7 214 4850 female 2009" ## [248] "Gentoo Biscoe 50.8 15.7 226 5200 male 2009" ## [249] "Gentoo Biscoe 49.4 15.8 216 4925 male 2009" ## [250] "Gentoo Biscoe 46.9 14.6 222 4875 female 2009" ## [251] "Gentoo Biscoe 48.4 14.4 203 4625 female 2009" ## [252] "Gentoo Biscoe 51.1 16.5 225 5250 male 2009" ## [253] "Gentoo Biscoe 48.5 15 219 4850 female 2009" ## [254] "Gentoo Biscoe 55.9 17 228 5600 male 2009" ## [255] "Gentoo Biscoe 47.2 15.5 215 4975 female 2009" ## [256] "Gentoo Biscoe 49.1 15 228 5500 male 2009" ## [257] "Gentoo Biscoe 47.3 13.8 216 4725 NA 2009" ## [258] "Gentoo Biscoe 46.8 16.1 215 5500 male 2009" ## [259] "Gentoo Biscoe 41.7 14.7 210 4700 female 2009" ## [260] "Gentoo Biscoe 53.4 15.8 219 5500 male 2009" ## [261] "Gentoo Biscoe 43.3 14 208 4575 female 2009" ## [262] "Gentoo Biscoe 48.1 15.1 209 5500 male 2009" ## [263] "Gentoo Biscoe 50.5 15.2 216 5000 female 2009" ## [264] "Gentoo Biscoe 49.8 15.9 229 5950 male 2009" ## [265] "Gentoo Biscoe 43.5 15.2 213 4650 female 2009" ## [266] "Gentoo Biscoe 51.5 16.3 230 5500 male 2009" ## [267] "Gentoo Biscoe 46.2 14.1 217 4375 female 2009" ## [268] "Gentoo Biscoe 55.1 16 230 5850 male 2009" ## [269] "Gentoo Biscoe 44.5 15.7 217 4875 NA 2009" ## [270] "Gentoo Biscoe 48.8 16.2 222 6000 male 2009" ## [271] "Gentoo Biscoe 47.2 13.7 214 4925 female 2009" ## [272] "Gentoo Biscoe NA NA NA NA NA 2009" ## [273] "Gentoo Biscoe 46.8 14.3 215 4850 female 2009" ## [274] "Gentoo Biscoe 50.4 15.7 222 5750 male 2009" ## [275] "Gentoo Biscoe 45.2 14.8 212 5200 female 2009" ## [276] "Gentoo Biscoe 49.9 16.1 213 5400 male 2009" ## [277] "Chinstrap Dream 46.5 17.9 192 3500 female 2007" ## [278] "Chinstrap Dream 50 19.5 196 3900 male 2007" ## [279] "Chinstrap Dream 51.3 19.2 193 3650 male 2007" ## [280] "Chinstrap Dream 45.4 18.7 188 3525 female 2007" ## [281] "Chinstrap Dream 52.7 19.8 197 3725 male 2007" ## [282] "Chinstrap Dream 45.2 17.8 198 3950 female 2007" ## [283] "Chinstrap Dream 46.1 18.2 178 3250 female 2007" ## [284] "Chinstrap Dream 51.3 18.2 197 3750 male 2007" ## [285] "Chinstrap Dream 46 18.9 195 4150 female 2007" ## [286] "Chinstrap Dream 51.3 19.9 198 3700 male 2007" ## [287] "Chinstrap Dream 46.6 17.8 193 3800 female 2007" ## [288] "Chinstrap Dream 51.7 20.3 194 3775 male 2007" ## [289] "Chinstrap Dream 47 17.3 185 3700 female 2007" ## [290] "Chinstrap Dream 52 18.1 201 4050 male 2007" ## [291] "Chinstrap Dream 45.9 17.1 190 3575 female 2007" ## [292] "Chinstrap Dream 50.5 19.6 201 4050 male 2007" ## [293] "Chinstrap Dream 50.3 20 197 3300 male 2007" ## [294] "Chinstrap Dream 58 17.8 181 3700 female 2007" ## [295] "Chinstrap Dream 46.4 18.6 190 3450 female 2007" ## [296] "Chinstrap Dream 49.2 18.2 195 4400 male 2007" ## [297] "Chinstrap Dream 42.4 17.3 181 3600 female 2007" ## [298] "Chinstrap Dream 48.5 17.5 191 3400 male 2007" ## [299] "Chinstrap Dream 43.2 16.6 187 2900 female 2007" ## [300] "Chinstrap Dream 50.6 19.4 193 3800 male 2007" ## [301] "Chinstrap Dream 46.7 17.9 195 3300 female 2007" ## [302] "Chinstrap Dream 52 19 197 4150 male 2007" ## [303] "Chinstrap Dream 50.5 18.4 200 3400 female 2008" ## [304] "Chinstrap Dream 49.5 19 200 3800 male 2008" ## [305] "Chinstrap Dream 46.4 17.8 191 3700 female 2008" ## [306] "Chinstrap Dream 52.8 20 205 4550 male 2008" ## [307] "Chinstrap Dream 40.9 16.6 187 3200 female 2008" ## [308] "Chinstrap Dream 54.2 20.8 201 4300 male 2008" ## [309] "Chinstrap Dream 42.5 16.7 187 3350 female 2008" ## [310] "Chinstrap Dream 51 18.8 203 4100 male 2008" ## [311] "Chinstrap Dream 49.7 18.6 195 3600 male 2008" ## [312] "Chinstrap Dream 47.5 16.8 199 3900 female 2008" ## [313] "Chinstrap Dream 47.6 18.3 195 3850 female 2008" ## [314] "Chinstrap Dream 52 20.7 210 4800 male 2008" ## [315] "Chinstrap Dream 46.9 16.6 192 2700 female 2008" ## [316] "Chinstrap Dream 53.5 19.9 205 4500 male 2008" ## [317] "Chinstrap Dream 49 19.5 210 3950 male 2008" ## [318] "Chinstrap Dream 46.2 17.5 187 3650 female 2008" ## [319] "Chinstrap Dream 50.9 19.1 196 3550 male 2008" ## [320] "Chinstrap Dream 45.5 17 196 3500 female 2008" ## [321] "Chinstrap Dream 50.9 17.9 196 3675 female 2009" ## [322] "Chinstrap Dream 50.8 18.5 201 4450 male 2009" ## [323] "Chinstrap Dream 50.1 17.9 190 3400 female 2009" ## [324] "Chinstrap Dream 49 19.6 212 4300 male 2009" ## [325] "Chinstrap Dream 51.5 18.7 187 3250 male 2009" ## [326] "Chinstrap Dream 49.8 17.3 198 3675 female 2009" ## [327] "Chinstrap Dream 48.1 16.4 199 3325 female 2009" ## [328] "Chinstrap Dream 51.4 19 201 3950 male 2009" ## [329] "Chinstrap Dream 45.7 17.3 193 3600 female 2009" ## [330] "Chinstrap Dream 50.7 19.7 203 4050 male 2009" ## [331] "Chinstrap Dream 42.5 17.3 187 3350 female 2009" ## [332] "Chinstrap Dream 52.2 18.8 197 3450 male 2009" ## [333] "Chinstrap Dream 45.2 16.6 191 3250 female 2009" ## [334] "Chinstrap Dream 49.3 19.9 203 4050 male 2009" ## [335] "Chinstrap Dream 50.2 18.8 202 3800 male 2009" ## [336] "Chinstrap Dream 45.6 19.4 194 3525 female 2009" ## [337] "Chinstrap Dream 51.9 19.5 206 3950 male 2009" ## [338] "Chinstrap Dream 46.8 16.5 189 3650 female 2009" ## [339] "Chinstrap Dream 45.7 17 195 3650 female 2009" ## [340] "Chinstrap Dream 55.8 19.8 207 4000 male 2009" ## [341] "Chinstrap Dream 43.5 18.1 202 3400 female 2009" ## [342] "Chinstrap Dream 49.6 18.2 193 3775 male 2009" ## [343] "Chinstrap Dream 50.8 19 210 4100 male 2009" ## [344] "Chinstrap Dream 50.2 18.7 198 3775 female 2009"Wrap up
Lots more to
{purrr}but we've covered a lotFunctional programming can really help your efficiency, and even if it slows you down initially, I'd recommend always striving toward it, because it will ultimately be a huge help.